New Arrival | μCaler Targeted Methylation Sequencing Drives New Breakthroughs in Multi-cance Early Screening (Part 2)
View: 5377 / Time: 2024-09-02
In part 1 (New Arrival | μCaler Targeted Methylation Sequencing Drives New Breakthroughs in Multi-cancer Early Screening (Part 1)), Nanodigmbio provided a comprehensive overview of the current status of early cancer screening and briefly introduced the upcoming μCaler Targeted Methylation Sequencing Comprehensive Solution. This end-to-end solution boasts several distinct features, including precise detection of large-scale CpG sites, accurate quantification of methylation levels, fast and convenient operation, highly efficient and stable, cost-effective sequencing, and stable delivery chain.
μCaler Targeted Methylation Sequencing Comprehensive Solution is an innovative solution developed by Nanodigmbio with independent intellectual property rights. Equipped with the exclusive patented μCaler Hybrid System, it aims to provide users with a new experience of accurate, stable, efficient, rapid, and simple methylation sequencing. In Part 2, we will showcase the outstanding performance of the μCaler Targeted Methylation Sequencing Comprehensive Solution through performance data and clinical sample application examples.
01 μCaler Targeted Methylation Sequencing Comprehensive Solution
1.1 Feature
Precise detection of large-scale CpG sites: Simultaneously assessment of multiple genes' CpG sites associated with various cancers, achieving precise comprehensive joint testing.
Accurate quantification of methylation levels: High-precision coverage for accurate quantification of different methylation states ranging from 0 to 100% in various samples.
Fast and convenient operation: Simplified experimental procedures, and user-friendly operations, completing the entire process within same day.
Highly efficient and stable: Avoid inconsistent amount of raw data, ensuring stability in capture sequencing results, thereby reducing the risk of rework.
Cost-effective sequencing: Utilizing a mixed hybrid mode to reduce the quantity of hybrid reaction, lowering sequencing costs and enhancing cost-effectiveness.
Stable delivery chain: In-house nucleic acid synthesis manufacturing ensures on-hand delivery and guarantees the stability of the delivery chain.
1.2 Features of μCaler Methylation Probe Design
μCaler Targeted Methylation Sequencing Comprehensive Solution utilizes a workflow of post-conversion capture. Its methylation probes are exhaustively designed based on the converted DNA template strands, achieving comprehensive coverage of targets with different methylation states. The main features include:
• Multiple probes rapidly search for targets to form stable binding, ensuring efficient capture of targets.
• Conjugation effect among multiple probes eliminate non-specific binding minimizing target loss.
• Probes capture information from both methylated and unmethylated states of double-stranded DNA.
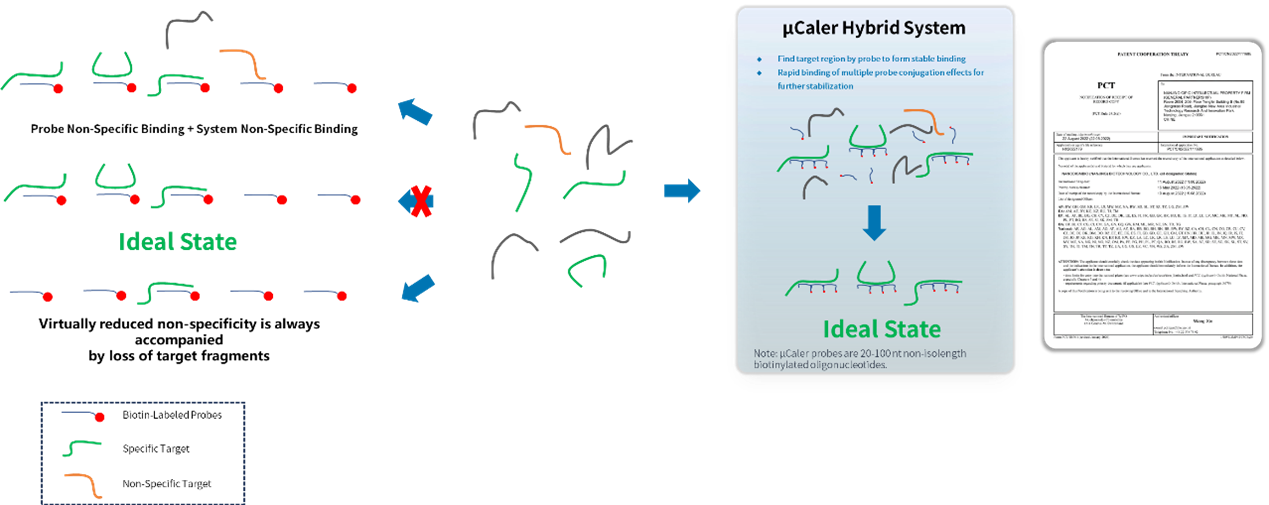
Figure 1. Design strategy of μCaler probe.
02 Performance
2.1 Precise Detection of Large-Scale CpG Sites
We have selected two customized panels with different scale CpG sites, based on NMPA-approved methylation detection kits and relevant methylation biomarkers reported in literature. The specific information is as follows:
Table 1. Basic information of custom panel.
Human genomic DNA standards (Promega, G1471) were used to prepare pre-libraries using the NadPrep Methyl Library Preparation Module coupled with the NadPrep Methyl Stubby Adapter (UDI) Module (with 10 nt Index) and NadPrep DNA Methyl Bisulfite Conversion Module. Hybrid capture were completed with μCaler Methyl Cervical Cancer Panel (MCC) and μCaler Methyl Multiple Cancer Panel (MMC). The results showed that μCaler Targeted Methylation Sequencing Comprehensive Solution achieves full coverage and highly stable reproducibility for CpG sites of different sizes. In addition, the on-target rate of the captured data was stably maintained at more than 40%, and the 0.2x mean CpG coverage also reached more than 99%. The solution offers high flexibility in customization, allowing users to choose different customized panels or expand without restrictions according to research needs to meet specific methylation site detection requirements.
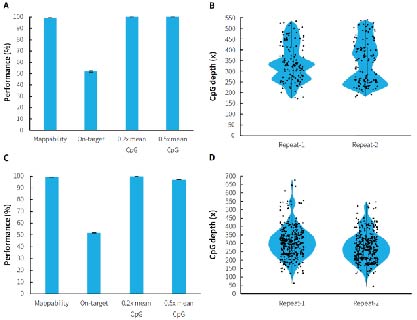
Figure 2. Performance of μCaler targeted methylation sequencing with custom panels containing different scales of CpG sites. A. Capture performance of MCC; B. CpG site coverage depth of MCC; C. Capture performance of MMC; D. CpG site coverage depth of MMC.
Note: Illumina® Novaseq 6000, PE 150 sequencing. For each sample, 1 Mb reads pair was randomly selected for data analysis.
2.2 Accurate Quantification of Methylation Levels
Applying μCaler Targeted Methylation Sequencing Comprehensive Solution to simulated samples with different methylation levels (0%, 10%, 50% and 100%) showed that this solution achieves high coverage depth and good uniformity in all tested samples (Figure 3. A). In addition, the methylation level detection results of the μCaler Targeted Methylation Sequencing Comprehensive Solution are highly consistent with the theoretical level (Figure 3. B), indicating reliable accuracy and repeatability. These results all show that μCaler Targeted Methylation Sequencing Comprehensive Solution is an efficient and reliable methylation detection technology that is suitable for research samples with various methylation levels.
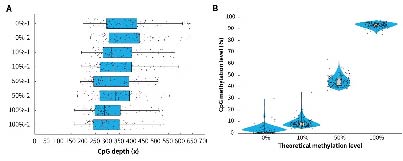
Figure 3. Performance of μCaler Targeted Methylation Sequencing with samples of different methylation levels. A. Coverage depth of CpG sites in samples with different methylation levels; B. Comparison of theoretical methylation levels of samples with the results of μCaler targeted methylation sequencing.
2.3 Fast and Convenient Operation
The traditional hybridization capture method has significant drawbacks, such as lengthy hybridization reaction time and complex, cumbersome experimental procedures. However, μCaler Hybrid System not only simplifies the experimental process of hybridization capture, but also further shortens the reaction time. As can be clearly seen from Figure 4., the entire process of the μCaler Hybrid System takes an average of only 4 hours, achieving high-speed and convenient operation. This enhancement not only speeds up the experiment, but also improves the laboratory efficiency, providing researchers with more convenient conditions and making hybridization capture technology more attractive in practical applications.
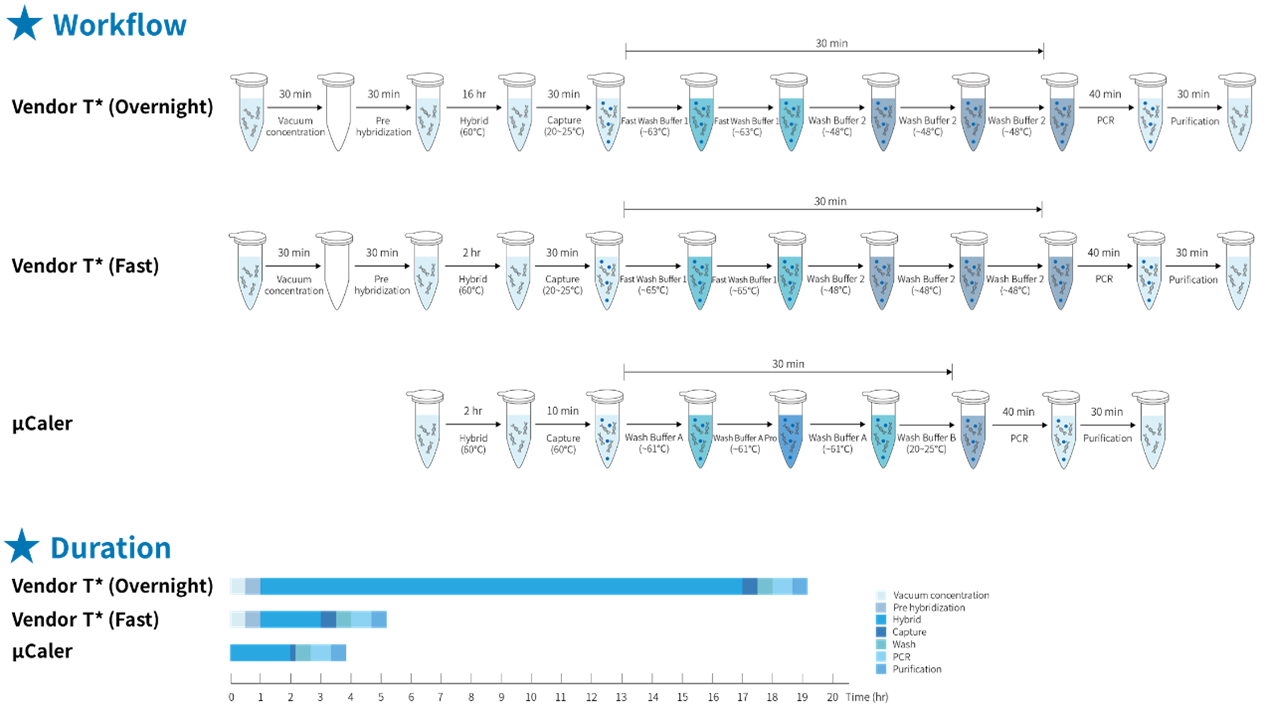
Figure 4. Comparison of experimental workflow and time consumption between traditional and μCaler hybridization capture.
2.4 Highly Efficient and Stable Capture
μCaler Hybrid System for Methylation has been optimized for special regions such as AT-rich, and when used with three customized mini panels, it still shows excellent capture performance and maintains high stable reproducibility. This optimized design makes μCaler Hybrid System for Methylation adaptable to different mini panels, providing users with more flexible and customizable options to meet different research needs.
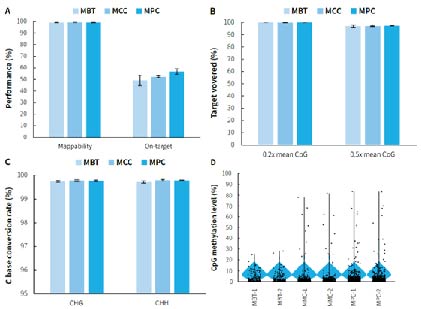
Figure 5. Capture performance of μCaler targeted methylation sequencing for mini panels. Pre-libraries were prepared using human genomic DNA standards (Promega, G1471), with 500 ng input per pre-library, and hybrid capture was completed with MBT, MMC, and MPC, respectively. A. Mappability and on-target rate; B. Target covered of CpG sites; C. Conversion rate of cytosine bases; D. CpG methylation levels.
Note: The coverage area for MBT is approximately 1.2 Kb, containing 159 CpG sites; while the coverage area for MPC is approximately 6.2 Kb, containing 620 CpG sites.
2.5 Cost-effective Sequencing
03 Application Example of Clinical Sample
Cancer early screening cohort studies employ various technical approaches to validate the methylation biomarker screened by the whole genome sequencing, aiming to meet diverse needs such as high sensitivity, wide coverage and controllable costs as much as possible. Researchers used μCaler targeted methylation sequencing (μCaler) and multiplex PCR amplification (Amp) to detect the methylation level of CpG sites in two clinical gDNA samples with different methylation levels (Tumor: relatively high methylation level; Normal: relatively low methylation level). Figure 7. shows that the detection results differ between the two technologies for samples with different methylation states. μCaler is suitable for the detection of samples with different methylation states, while Amp has a certain preference for targets with different methylation states, and some areas cannot be covered. Further results for CpG sites related to chromosome 7 (Chr 7) show that the methylation level detected by Amp for samples with high methylation status is lower than that detected by μCaler, while the detection result for samples with low methylation status is N.
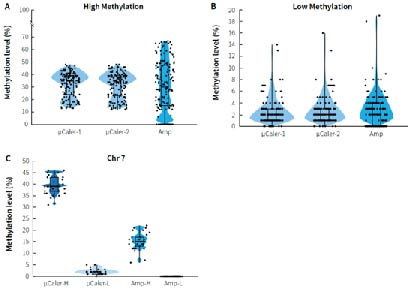
Figure 7. Comparison of detection results from different technical approaches using clinical samples. A. Detection results for samples with high methylation levels (Tumor); B. Detection results for samples with low methylation levels (Normal); C. Methylation levels of CpG sites related to chromosome 7 (Chr 7).
Note: μCaler targeted methylation sequencing was performed with MBT for hybrid capture.
The reasons for these differences are mainly as follows: 1) Designing primers for multiplex PCR amplification is highly complex, and the amplification process has certain biases for regions with different methylation states; 2) After conversion, GC-rich regions will become AT-rich or remain GC-rich, which makes PCR amplification more difficult. This can lead to poor coverage or even no coverage in some regions; 3) When designing primers for methylation-specific PCR, the primer binding site cannot contain CpG sites. This makes it impossible to design primers for regions with consecutive CpG regions.
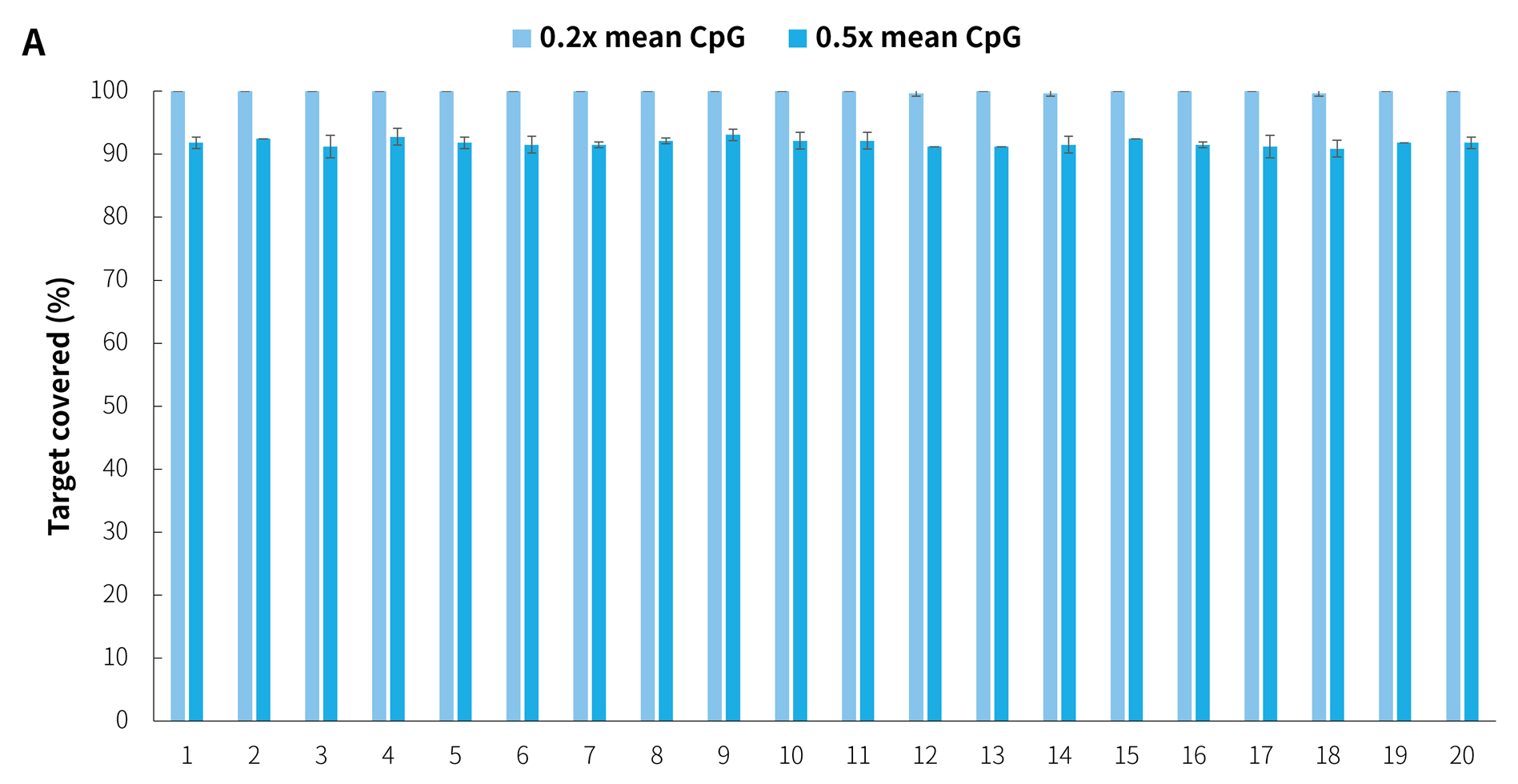
In addition, in the detection of 20 clinical samples, μCaler Hybrid System for Methylation still showed stable capture performance, as shown in Figure 8.
04 Application Prospects
The cancer early screening industry presents a differentiated competitive landscape, with single-cancer screening mainly relying on PCR technology and multi-cancer screening primarily uses NGS technology, which presents relatively higher technical barriers. In this context, Nanodigmbio can provide users with a free online probe design platform, allowing users to flexibly design probes according to their personalized needs and providing customized product solutions. Among them, μCaler hybrid capture technology not only has independent intellectual property protection, but also provides a solid foundation for the successful implementation of products, ensuring robust support for client’s development in the field of cancer early screening.
Clinical research on early screening products is mainly divided into retrospective and prospective studies. Nanodigmbio focuses on providing end-to-end solutions and technical support for clinical cohort studies, including methylation site screening, verification and early screening product development. Comprehensive technical solutions will help accelerate the research process of early screening products, contributing to the field of cancer early diagnosis.
μCaler Targeted Methylation Sequencing Comprehensive Solution is an innovative solution developed by Nanodigmbio with independent intellectual property rights. Equipped with the exclusive patented μCaler Hybrid System, it aims to provide users with a new experience of accurate, stable, efficient, rapid, and simple methylation sequencing. In Part 2, we will showcase the outstanding performance of the μCaler Targeted Methylation Sequencing Comprehensive Solution through performance data and clinical sample application examples.
01 μCaler Targeted Methylation Sequencing Comprehensive Solution
1.1 Feature
Precise detection of large-scale CpG sites: Simultaneously assessment of multiple genes' CpG sites associated with various cancers, achieving precise comprehensive joint testing.
Accurate quantification of methylation levels: High-precision coverage for accurate quantification of different methylation states ranging from 0 to 100% in various samples.
Fast and convenient operation: Simplified experimental procedures, and user-friendly operations, completing the entire process within same day.
Highly efficient and stable: Avoid inconsistent amount of raw data, ensuring stability in capture sequencing results, thereby reducing the risk of rework.
Cost-effective sequencing: Utilizing a mixed hybrid mode to reduce the quantity of hybrid reaction, lowering sequencing costs and enhancing cost-effectiveness.
Stable delivery chain: In-house nucleic acid synthesis manufacturing ensures on-hand delivery and guarantees the stability of the delivery chain.
1.2 Features of μCaler Methylation Probe Design
μCaler Targeted Methylation Sequencing Comprehensive Solution utilizes a workflow of post-conversion capture. Its methylation probes are exhaustively designed based on the converted DNA template strands, achieving comprehensive coverage of targets with different methylation states. The main features include:
• Multiple probes rapidly search for targets to form stable binding, ensuring efficient capture of targets.
• Conjugation effect among multiple probes eliminate non-specific binding minimizing target loss.
• Probes capture information from both methylated and unmethylated states of double-stranded DNA.

Figure 1. Design strategy of μCaler probe.
02 Performance
2.1 Precise Detection of Large-Scale CpG Sites
We have selected two customized panels with different scale CpG sites, based on NMPA-approved methylation detection kits and relevant methylation biomarkers reported in literature. The specific information is as follows:
Table 1. Basic information of custom panel.
Human genomic DNA standards (Promega, G1471) were used to prepare pre-libraries using the NadPrep Methyl Library Preparation Module coupled with the NadPrep Methyl Stubby Adapter (UDI) Module (with 10 nt Index) and NadPrep DNA Methyl Bisulfite Conversion Module. Hybrid capture were completed with μCaler Methyl Cervical Cancer Panel (MCC) and μCaler Methyl Multiple Cancer Panel (MMC). The results showed that μCaler Targeted Methylation Sequencing Comprehensive Solution achieves full coverage and highly stable reproducibility for CpG sites of different sizes. In addition, the on-target rate of the captured data was stably maintained at more than 40%, and the 0.2x mean CpG coverage also reached more than 99%. The solution offers high flexibility in customization, allowing users to choose different customized panels or expand without restrictions according to research needs to meet specific methylation site detection requirements.

Figure 2. Performance of μCaler targeted methylation sequencing with custom panels containing different scales of CpG sites. A. Capture performance of MCC; B. CpG site coverage depth of MCC; C. Capture performance of MMC; D. CpG site coverage depth of MMC.
Note: Illumina® Novaseq 6000, PE 150 sequencing. For each sample, 1 Mb reads pair was randomly selected for data analysis.
2.2 Accurate Quantification of Methylation Levels
Applying μCaler Targeted Methylation Sequencing Comprehensive Solution to simulated samples with different methylation levels (0%, 10%, 50% and 100%) showed that this solution achieves high coverage depth and good uniformity in all tested samples (Figure 3. A). In addition, the methylation level detection results of the μCaler Targeted Methylation Sequencing Comprehensive Solution are highly consistent with the theoretical level (Figure 3. B), indicating reliable accuracy and repeatability. These results all show that μCaler Targeted Methylation Sequencing Comprehensive Solution is an efficient and reliable methylation detection technology that is suitable for research samples with various methylation levels.

Figure 3. Performance of μCaler Targeted Methylation Sequencing with samples of different methylation levels. A. Coverage depth of CpG sites in samples with different methylation levels; B. Comparison of theoretical methylation levels of samples with the results of μCaler targeted methylation sequencing.
2.3 Fast and Convenient Operation
The traditional hybridization capture method has significant drawbacks, such as lengthy hybridization reaction time and complex, cumbersome experimental procedures. However, μCaler Hybrid System not only simplifies the experimental process of hybridization capture, but also further shortens the reaction time. As can be clearly seen from Figure 4., the entire process of the μCaler Hybrid System takes an average of only 4 hours, achieving high-speed and convenient operation. This enhancement not only speeds up the experiment, but also improves the laboratory efficiency, providing researchers with more convenient conditions and making hybridization capture technology more attractive in practical applications.

Figure 4. Comparison of experimental workflow and time consumption between traditional and μCaler hybridization capture.
2.4 Highly Efficient and Stable Capture
μCaler Hybrid System for Methylation has been optimized for special regions such as AT-rich, and when used with three customized mini panels, it still shows excellent capture performance and maintains high stable reproducibility. This optimized design makes μCaler Hybrid System for Methylation adaptable to different mini panels, providing users with more flexible and customizable options to meet different research needs.

Figure 5. Capture performance of μCaler targeted methylation sequencing for mini panels. Pre-libraries were prepared using human genomic DNA standards (Promega, G1471), with 500 ng input per pre-library, and hybrid capture was completed with MBT, MMC, and MPC, respectively. A. Mappability and on-target rate; B. Target covered of CpG sites; C. Conversion rate of cytosine bases; D. CpG methylation levels.
Note: The coverage area for MBT is approximately 1.2 Kb, containing 159 CpG sites; while the coverage area for MPC is approximately 6.2 Kb, containing 620 CpG sites.
2.5 Cost-effective Sequencing
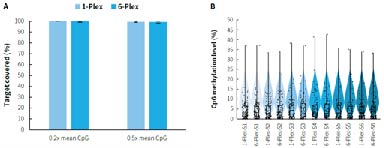
Targeted methylation sequencing can be costly. By establishing a multi-sample mixed hybridization scheme, the cost of reagents and labor is effectively reduced, significantly improving experimental efficiency. As shown in Figure 6., compared to single library hybridization,μCaler Hybrid System for Methylation using mixed hybridization of 6 libraries not only lowers experimental costs but also maintains data quality, achieving a higher price-performance ratio.
03 Application Example of Clinical Sample
Cancer early screening cohort studies employ various technical approaches to validate the methylation biomarker screened by the whole genome sequencing, aiming to meet diverse needs such as high sensitivity, wide coverage and controllable costs as much as possible. Researchers used μCaler targeted methylation sequencing (μCaler) and multiplex PCR amplification (Amp) to detect the methylation level of CpG sites in two clinical gDNA samples with different methylation levels (Tumor: relatively high methylation level; Normal: relatively low methylation level). Figure 7. shows that the detection results differ between the two technologies for samples with different methylation states. μCaler is suitable for the detection of samples with different methylation states, while Amp has a certain preference for targets with different methylation states, and some areas cannot be covered. Further results for CpG sites related to chromosome 7 (Chr 7) show that the methylation level detected by Amp for samples with high methylation status is lower than that detected by μCaler, while the detection result for samples with low methylation status is N.

Figure 7. Comparison of detection results from different technical approaches using clinical samples. A. Detection results for samples with high methylation levels (Tumor); B. Detection results for samples with low methylation levels (Normal); C. Methylation levels of CpG sites related to chromosome 7 (Chr 7).
Note: μCaler targeted methylation sequencing was performed with MBT for hybrid capture.
The reasons for these differences are mainly as follows: 1) Designing primers for multiplex PCR amplification is highly complex, and the amplification process has certain biases for regions with different methylation states; 2) After conversion, GC-rich regions will become AT-rich or remain GC-rich, which makes PCR amplification more difficult. This can lead to poor coverage or even no coverage in some regions; 3) When designing primers for methylation-specific PCR, the primer binding site cannot contain CpG sites. This makes it impossible to design primers for regions with consecutive CpG regions.
In addition, in the detection of 20 clinical samples, μCaler Hybrid System for Methylation still showed stable capture performance, as shown in Figure 8.

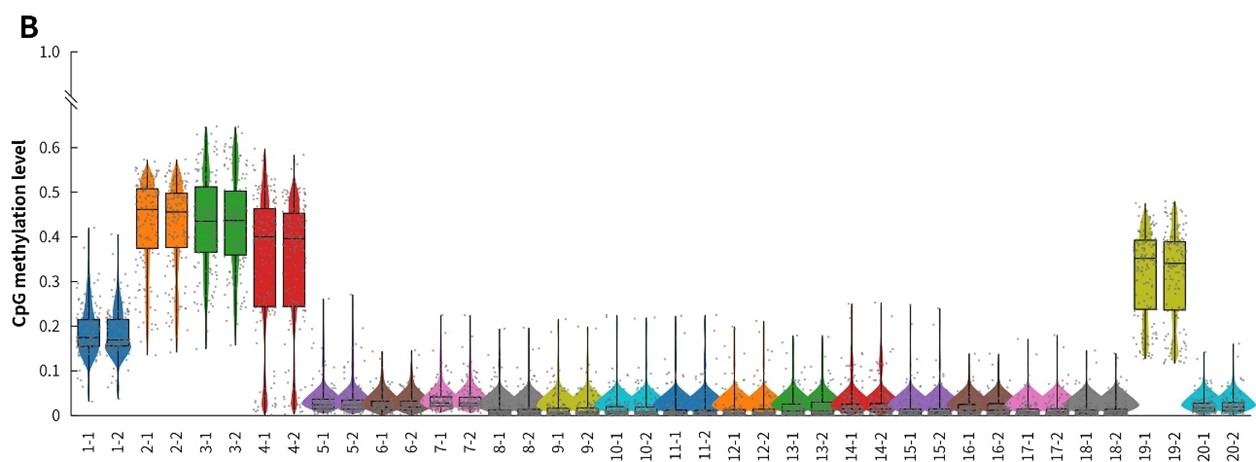
Figure 8. Capture performance of μCaler targeted methylation sequencing with clinical samples. A. CpG site coverage in the target region; B. Methylation level of CpG sites.
In summary, compared with multiplex PCR amplification technology, μCaler Targeted Methylation Sequencing Comprehensive Solution shows higher detection sensitivity and stability in the detection of clinical samples.04 Application Prospects
The cancer early screening industry presents a differentiated competitive landscape, with single-cancer screening mainly relying on PCR technology and multi-cancer screening primarily uses NGS technology, which presents relatively higher technical barriers. In this context, Nanodigmbio can provide users with a free online probe design platform, allowing users to flexibly design probes according to their personalized needs and providing customized product solutions. Among them, μCaler hybrid capture technology not only has independent intellectual property protection, but also provides a solid foundation for the successful implementation of products, ensuring robust support for client’s development in the field of cancer early screening.
Clinical research on early screening products is mainly divided into retrospective and prospective studies. Nanodigmbio focuses on providing end-to-end solutions and technical support for clinical cohort studies, including methylation site screening, verification and early screening product development. Comprehensive technical solutions will help accelerate the research process of early screening products, contributing to the field of cancer early diagnosis.
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)



