New Arrival | μCaler Targeted Methylation Sequencing Drives New Breakthroughs in Multi-cancer Early Screening (Part 1)
View: 5331 / Time: 2024-09-02
01 Current Status of Early Cancer Screening
1.1 Early screening is a crucial method for improving the survival rate of cancer patients
1.2 DNA methylation is a "promising candidate biomarker" for early cancer screening.
There are many potential biomarkers that can be used for early cancer screening clinically, including proteins, fragmentomics, small molecule metabolites, miRNA, ctDNA (circulating tumor DNA) mutations, and ctDNA methylation. Among these, DNA methylation occurs in almost all precancerous lesions and at the early stages of cancer, making it one of the most promising early cancer screening biomarker. ctDNA methylation, as an early cancer screening marker, has the following advantages: high and stable signal abundance; strong organ specificity, enabling tissue tracing; clustered distribution across the genome, resulting in a higher signal-to-noise ratio.
There are several technical approaches for DNA methylation detection, such as whole genome bisulfite sequencing (WGBS), reduced representation bisulfite sequencing (RRBS), 450/850K microarrays, targeted methylation sequencing using liquid phase hybridization capture technology (hybridization capture sequencing), multiplex PCR amplification sequencing, pyrosequencing, mass spectrometry, and methylation-specific PCR (MSP), etc. Currently, most products for early screening based on methylation primarily utilize the technical approaches of MSP, multiplex PCR amplification sequencing, and hybridization capture sequencing (Table 1.).
Tabel 1. Technical approaches of methylation early screening products.
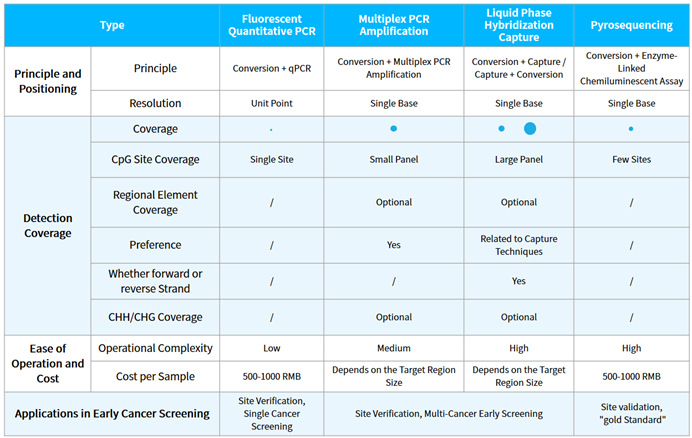
NMPA has approved 19 methylation detection products for single cancer type, all of which use the MSP method (until June, 2023). This method is only suitable for qualitative detection of the methylation statuses of a limited number of genes. It is simple, convenient and cost-effective. However, it also faces challenges in primer design, leading to potential false-positive results.
Multiplex PCR amplification sequencing and hybrid capture sequencing can simultaneously detect the methylation status of multiple CpG sites, making them more suitable for multi-cancer early screening. However, the challenge with multiplex PCR amplification sequencing lies in the difficulty of primer design, especially for consecutive CpG sites. Hybrid capture sequencing is suitable for capturing targets with different methylation statuses, and it has more potential application scenario
02 μCaler Targeted Methylation Sequencing Comprehensive Solution
2.1 Introduction
μCaler Targeted Methylation Sequencing Comprehensive Solution, which featuring an exclusive patented μCaler hybridization capture system. This independently innovative solution aims to provide users with a new experience in methylation sequencing that is accurate, stable, efficient, rapid, and simple.
Exclusive patented μCaler technology: This solution features the globally innovative μCaler Hybrid System, incorporating an optimized hybrid capture system and a patented probe design scheme. It ensures efficient capture of converted methylation libraries and accurate detection of different methylation states. The application of this patented technology provides users with more comprehensive and accurate methylation information.
Optimization for specific regions: This solution with a hybridization capture system optimized for methylation is not only suitable for stable capture with mini-Panels but also applicable for capturing special regions like AT-rich sequences. It enables a more comprehensive analysis of complex genomic regions.
Entire process within same day: This solution offers both stability and efficiency, allowing the entire process of methylated captured library to be completed within same day. This provides users with exceptional convenience and an unprecedented efficient experience.
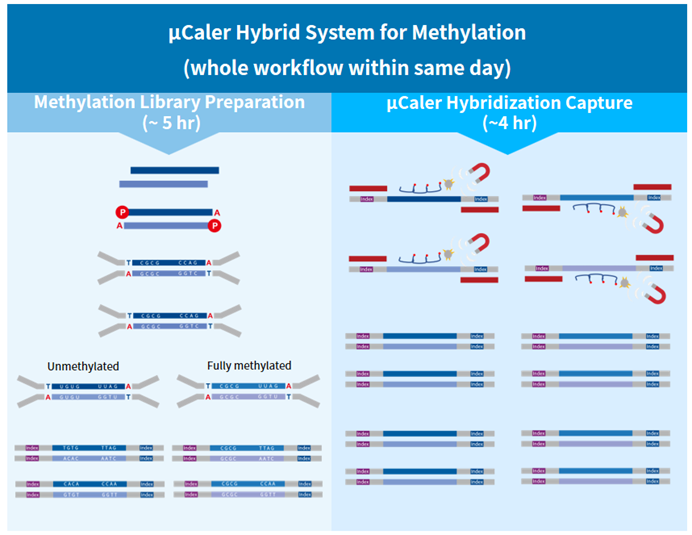
Figure 3. Workflow of μCaler Targeted Methylation Sequencing Comprehensive Solution.
2.2 Features
Precise detection of large-scale CpG sites: Simultaneously assessment of multiple genes' CpG sites associated with various cancers, achieving precise comprehensive joint testing.
Accurate quantification of methylation levels: High-precision coverage for accurate quantification of different methylation states ranging from 0 to 100% in various samples.
Fast and convenient operation: Simplified experimental procedures, and user-friendly operations, completing the entire process within same day.
Highly Efficient and Stable: Avoid inconsistent amount of raw data, ensuring stability in capture sequencing results, thereby reducing the risk of rework.
Cost-effective sequencing: Utilizing a mixed hybrid mode to reduce the quantity of hybrid reaction, lowering sequencing costs and enhancing cost-effectiveness.
Stable delivery chain: In-house nucleic acid synthesis manufacturing ensures on-hand delivery and guarantees the stability of the delivery chain.
2.3 Performance
2.3.1 Highly Efficient and Stable
Using the μCaler Hybrid System for Methylation, the capture results for varying hybridization input amounts of a single library (500 ng - 3,000 ng), show that the mappability of the captured data consistently remains above 99%, and the on-target rate stay above 50%. Additionally, the uniformity of CpG site coverage is excellent, with a 0.2x mean CpG coverage more than 99%. This indicates that the μCaler Hybrid System for Methylation supports captured library inputs ranging from 500 ng to 3,000 ng.
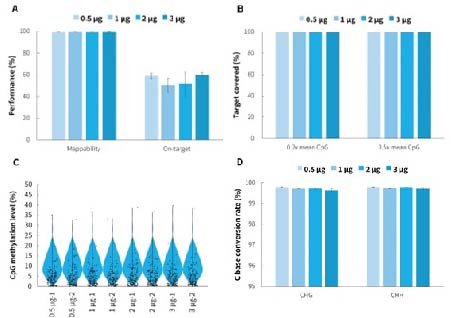
Figure 4. Capture performance of μCaler Targeted Methylation Sequencing Comprehensive Solution. Human Genomic DNA (Promega, G1471) were used to prepare prr-libraries with the NadPrep Methyl Library Preparation Module coupled with the NadPrep Methyl Stubby Adapter (UDI) Module (with 10 nt Index) and NadPrep DNA Methyl Bisulfite Conversion Module. Hybrid capture were completed withμCaler Methyl Cervical Cancer Pane (MCC)l. A. Mappability & On-target rate. B. Uniformity of CpG site coverage. C. Methylation level of CpG sites. D. Conversion rate of C bases.
Note: The coverage area for MCC is approximately 1.4 Kb, containing 125 CpG sites.
2.3.2 Accurate Quantification of Methylation Levels
Pyrosequencing (Pyro) utilizes an enzyme cascade chemiluminescent reaction to rapidly detect CpG site methylation levels, making it the "gold standard" for methylation detection. Therefore, it can be used for accurate comparison and validation of the precision of μCaler Targeted Methylation Sequencing.
The Human HCT116 DKO Methylated DNA (Zymo, D5014-2) as a positive control and Human HCT116 DKO Non-Methylated DNA (Zymo, D5014-1) as a negative control were mixed in different proportions to simulate different methylation levels. Subsequently, we used μCaler Targeted Methylation Sequencing (μCaler) and pyrosequencing (Pyro) to detect the methylation levels of multiple CpG sites in multiple genes (38 CpG sites in genes MGMT, Septin9, and BMP3) in the simulated samples. The results showed significant deviations in the detecting results of Pyro for the samples with 0%, 2%, and 100% methylation levels exhibited. In contrast, μCaler provided more accurate detection across different methylation levels. This indicates that μCaler can accurately reflect the true methylation status of samples and can be widely applied in epigenetic research.
Pyro uses the converted products as templates for amplification. After conversion, CpG sites become AT-rich or GC-rich regions. Both the amplification and sequencing primers exhibit preferences for templates with different GC content, leading to deviations from the true methylation levels in samples with low or high methylation levels. In contrast; μCaler designs probes targeting the converted templates using an exhaustive probe design approach. This allows for the capture of all methylation states, enabling more accurate detection of samples with different methylation levels through universal primer amplification and sequencing of the library.
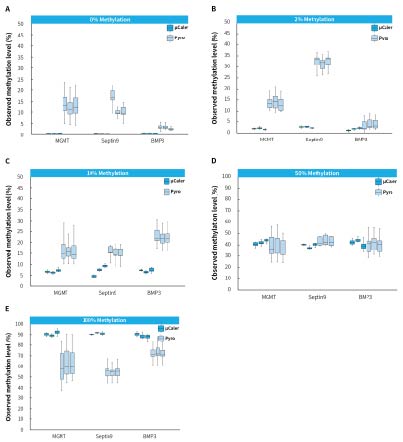
Figure 5. Methylation levels detected by different methods in simulated samples. A-E. represent simulated samples with methylation levels of 0%, 2%, 10%, 50%, and 100%, respectively. The input amount for pyrosequencing is 200 ng, while for μCaler Targeted Methylation Sequencing is 25 ng.
Reference
[1] Xia C, Basu P, Kramer B S, et al. Cancer screening in China: a steep road from evidence to implementation[J]. The Lancet Public Health, 2023.
[2] Robertson K D. DNA methylation and human disease[J]. Nature Reviews Genetics, 2005, 6(8): 597-610.
[3] Oxnard G R, Klein E A, Seiden M V, et al. Simultaneous multi-cancer detection and tissue of origin (TOO) localization using targeted bisulfite sequencing of plasma cell-free DNA (cfDNA)[J]. Annals of Oncology, 2019, 30: v912.
1.1 Early screening is a crucial method for improving the survival rate of cancer patients
According to data from IARC, 19.29 million new cancer cases globally occurred in 2021, with about 9.96 million deaths. The global cancer burden is expected to reach 28.4 million cases in 2040, reflecting a 47% increase from 2020. Additionally, the five-year survival rates of cancer patients vary among different countries. This is due to the severe lack of cancer screening in China, with most patients being diagnosed at middle or late stages, leading to poor treatment effect (Figure 1.).
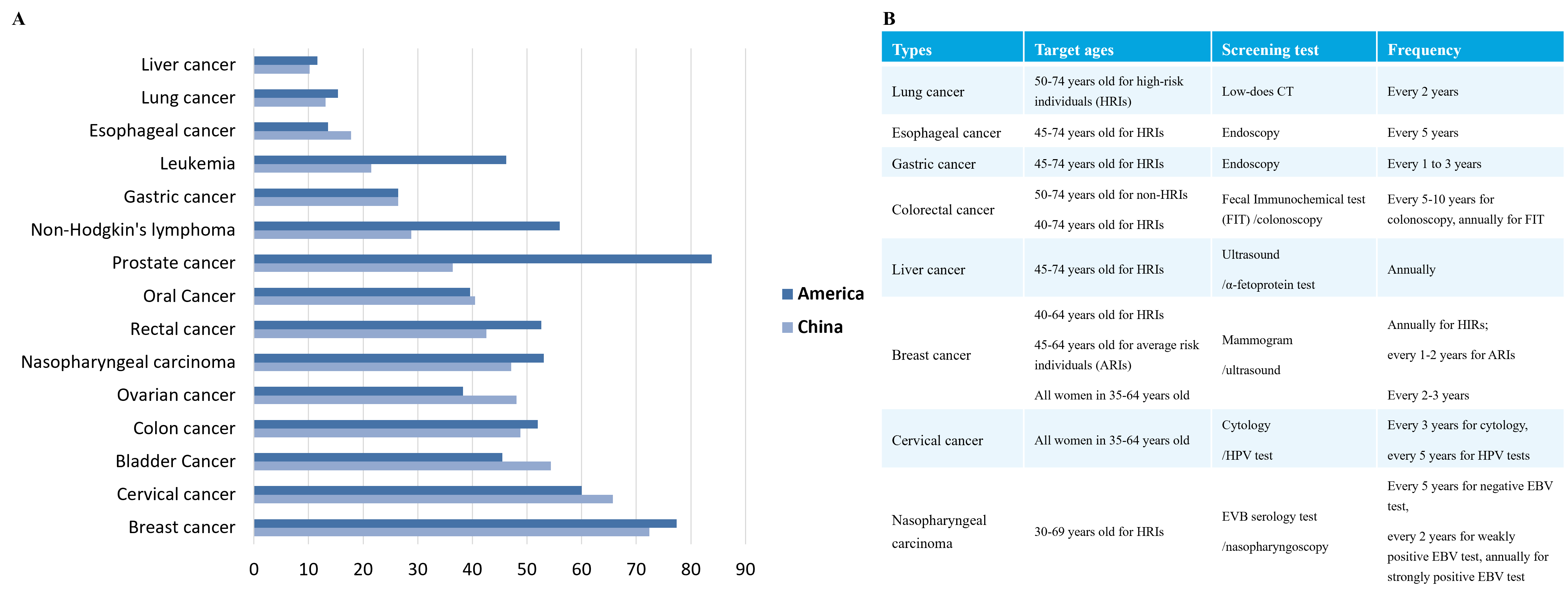
1.2 DNA methylation is a "promising candidate biomarker" for early cancer screening.
There are many potential biomarkers that can be used for early cancer screening clinically, including proteins, fragmentomics, small molecule metabolites, miRNA, ctDNA (circulating tumor DNA) mutations, and ctDNA methylation. Among these, DNA methylation occurs in almost all precancerous lesions and at the early stages of cancer, making it one of the most promising early cancer screening biomarker. ctDNA methylation, as an early cancer screening marker, has the following advantages: high and stable signal abundance; strong organ specificity, enabling tissue tracing; clustered distribution across the genome, resulting in a higher signal-to-noise ratio.
There are several technical approaches for DNA methylation detection, such as whole genome bisulfite sequencing (WGBS), reduced representation bisulfite sequencing (RRBS), 450/850K microarrays, targeted methylation sequencing using liquid phase hybridization capture technology (hybridization capture sequencing), multiplex PCR amplification sequencing, pyrosequencing, mass spectrometry, and methylation-specific PCR (MSP), etc. Currently, most products for early screening based on methylation primarily utilize the technical approaches of MSP, multiplex PCR amplification sequencing, and hybridization capture sequencing (Table 1.).
Tabel 1. Technical approaches of methylation early screening products.

NMPA has approved 19 methylation detection products for single cancer type, all of which use the MSP method (until June, 2023). This method is only suitable for qualitative detection of the methylation statuses of a limited number of genes. It is simple, convenient and cost-effective. However, it also faces challenges in primer design, leading to potential false-positive results.
Multiplex PCR amplification sequencing and hybrid capture sequencing can simultaneously detect the methylation status of multiple CpG sites, making them more suitable for multi-cancer early screening. However, the challenge with multiplex PCR amplification sequencing lies in the difficulty of primer design, especially for consecutive CpG sites. Hybrid capture sequencing is suitable for capturing targets with different methylation statuses, and it has more potential application scenario
02 μCaler Targeted Methylation Sequencing Comprehensive Solution
2.1 Introduction
μCaler Targeted Methylation Sequencing Comprehensive Solution, which featuring an exclusive patented μCaler hybridization capture system. This independently innovative solution aims to provide users with a new experience in methylation sequencing that is accurate, stable, efficient, rapid, and simple.
Exclusive patented μCaler technology: This solution features the globally innovative μCaler Hybrid System, incorporating an optimized hybrid capture system and a patented probe design scheme. It ensures efficient capture of converted methylation libraries and accurate detection of different methylation states. The application of this patented technology provides users with more comprehensive and accurate methylation information.
Optimization for specific regions: This solution with a hybridization capture system optimized for methylation is not only suitable for stable capture with mini-Panels but also applicable for capturing special regions like AT-rich sequences. It enables a more comprehensive analysis of complex genomic regions.
Entire process within same day: This solution offers both stability and efficiency, allowing the entire process of methylated captured library to be completed within same day. This provides users with exceptional convenience and an unprecedented efficient experience.

Figure 3. Workflow of μCaler Targeted Methylation Sequencing Comprehensive Solution.
2.2 Features
Precise detection of large-scale CpG sites: Simultaneously assessment of multiple genes' CpG sites associated with various cancers, achieving precise comprehensive joint testing.
Accurate quantification of methylation levels: High-precision coverage for accurate quantification of different methylation states ranging from 0 to 100% in various samples.
Fast and convenient operation: Simplified experimental procedures, and user-friendly operations, completing the entire process within same day.
Highly Efficient and Stable: Avoid inconsistent amount of raw data, ensuring stability in capture sequencing results, thereby reducing the risk of rework.
Cost-effective sequencing: Utilizing a mixed hybrid mode to reduce the quantity of hybrid reaction, lowering sequencing costs and enhancing cost-effectiveness.
Stable delivery chain: In-house nucleic acid synthesis manufacturing ensures on-hand delivery and guarantees the stability of the delivery chain.
2.3 Performance
2.3.1 Highly Efficient and Stable
Using the μCaler Hybrid System for Methylation, the capture results for varying hybridization input amounts of a single library (500 ng - 3,000 ng), show that the mappability of the captured data consistently remains above 99%, and the on-target rate stay above 50%. Additionally, the uniformity of CpG site coverage is excellent, with a 0.2x mean CpG coverage more than 99%. This indicates that the μCaler Hybrid System for Methylation supports captured library inputs ranging from 500 ng to 3,000 ng.

Figure 4. Capture performance of μCaler Targeted Methylation Sequencing Comprehensive Solution. Human Genomic DNA (Promega, G1471) were used to prepare prr-libraries with the NadPrep Methyl Library Preparation Module coupled with the NadPrep Methyl Stubby Adapter (UDI) Module (with 10 nt Index) and NadPrep DNA Methyl Bisulfite Conversion Module. Hybrid capture were completed withμCaler Methyl Cervical Cancer Pane (MCC)l. A. Mappability & On-target rate. B. Uniformity of CpG site coverage. C. Methylation level of CpG sites. D. Conversion rate of C bases.
Note: The coverage area for MCC is approximately 1.4 Kb, containing 125 CpG sites.
2.3.2 Accurate Quantification of Methylation Levels
Pyrosequencing (Pyro) utilizes an enzyme cascade chemiluminescent reaction to rapidly detect CpG site methylation levels, making it the "gold standard" for methylation detection. Therefore, it can be used for accurate comparison and validation of the precision of μCaler Targeted Methylation Sequencing.
The Human HCT116 DKO Methylated DNA (Zymo, D5014-2) as a positive control and Human HCT116 DKO Non-Methylated DNA (Zymo, D5014-1) as a negative control were mixed in different proportions to simulate different methylation levels. Subsequently, we used μCaler Targeted Methylation Sequencing (μCaler) and pyrosequencing (Pyro) to detect the methylation levels of multiple CpG sites in multiple genes (38 CpG sites in genes MGMT, Septin9, and BMP3) in the simulated samples. The results showed significant deviations in the detecting results of Pyro for the samples with 0%, 2%, and 100% methylation levels exhibited. In contrast, μCaler provided more accurate detection across different methylation levels. This indicates that μCaler can accurately reflect the true methylation status of samples and can be widely applied in epigenetic research.
Pyro uses the converted products as templates for amplification. After conversion, CpG sites become AT-rich or GC-rich regions. Both the amplification and sequencing primers exhibit preferences for templates with different GC content, leading to deviations from the true methylation levels in samples with low or high methylation levels. In contrast; μCaler designs probes targeting the converted templates using an exhaustive probe design approach. This allows for the capture of all methylation states, enabling more accurate detection of samples with different methylation levels through universal primer amplification and sequencing of the library.

Figure 5. Methylation levels detected by different methods in simulated samples. A-E. represent simulated samples with methylation levels of 0%, 2%, 10%, 50%, and 100%, respectively. The input amount for pyrosequencing is 200 ng, while for μCaler Targeted Methylation Sequencing is 25 ng.
We will showcase more performance and real sample application examples in the next part. Stay tuned!
[1] Xia C, Basu P, Kramer B S, et al. Cancer screening in China: a steep road from evidence to implementation[J]. The Lancet Public Health, 2023.
[2] Robertson K D. DNA methylation and human disease[J]. Nature Reviews Genetics, 2005, 6(8): 597-610.
[3] Oxnard G R, Klein E A, Seiden M V, et al. Simultaneous multi-cancer detection and tissue of origin (TOO) localization using targeted bisulfite sequencing of plasma cell-free DNA (cfDNA)[J]. Annals of Oncology, 2019, 30: v912.
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)



