Product details
NadPrep Single Cell WGA Kit
#1300102
NadPrep Single Cell WGA Kit is designed for whole-genome amplification (WGA) of genomic DNA (gDNA) from single cells or ultra-low amounts of purified gDNA. Utilizing optimized buffers and enzymes, whole-genome amplification can be completed within 3 hr. The kit features a simple, streamlined workflow including cell lysis, preamplification, and amplification to generate microgram-level gDNA with high coverage and uniformity.
- Product details
- Kit Content
- FAQ
- Ordering Information
- Resources
Feature
● Designed for single/multiple cells (1-10) or ultra-low gDNA (> 30 pg) inputs● Enhances sequencing coverage and uniformity, thereby improving accuracy of variants detection
● Broadly compatible with downstream applications including WGS, large fragment CNVs analysis, qPCR analysis, etc.
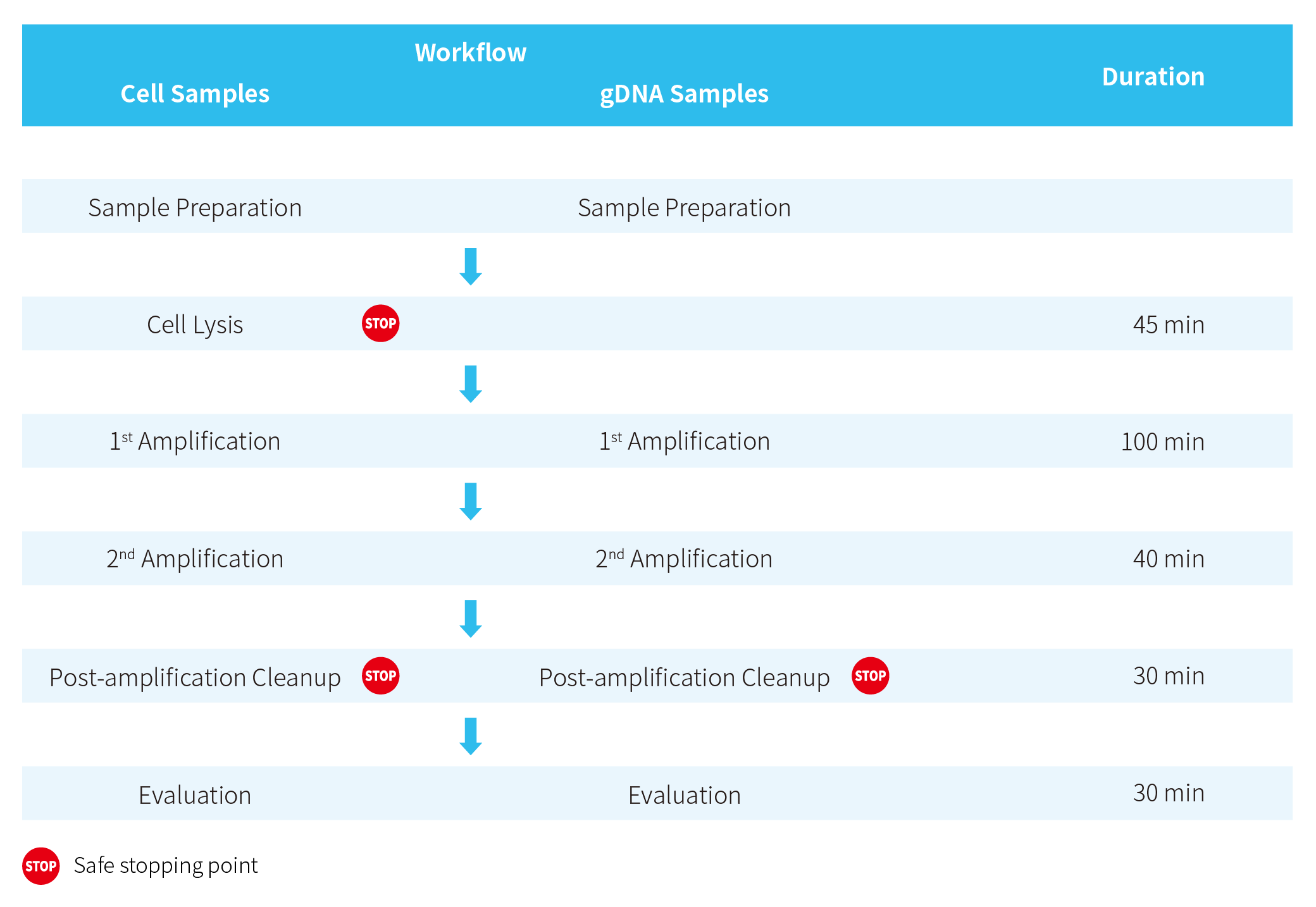
Workflow
Performance
High Concordance in CNVs Analysis
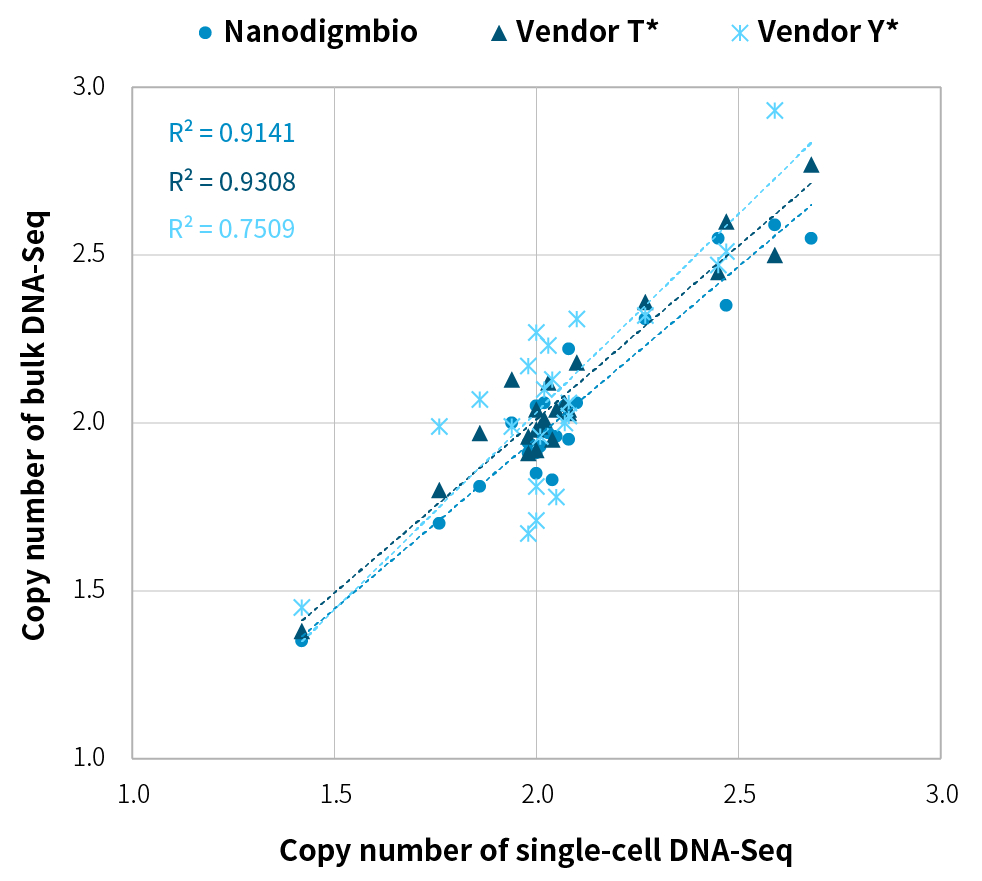
Figure 1. Comparison of CNVs analysis results between single-cell DNA-Seq and bulk DNA-Seq in 293T cells. The correlation of average copy numbers across different libraries demonstrates high concordance between single-cell DNA-Seq from Nanodigmbio’s solution and bulk DNA-Seq (R2 = 0.9141). Single-cell DNA-Seq libraries were prepared from 5-10 cells amplified with different WGA kits (Nanodigmbio, Vendor T* and Vendor Y*). 100 ng of each amplified DNA were used for library preparation with NadPrep EZ DNA Library Preparation Module v2 and NadPrep Universal Stubby Adapter (UDI) Module. Bulk DNA-Seq libraries were prepared from 100 ng gDNA extracted from cell lines. Libraries were sequenced on Illumina NovaSeq 6000 (PE150), generating approximately 1 Gb of raw data per sample. Variant analysis was performed with CNVkit in 1,000-kb bin sizes.
Note: Samples were cultured 293T cells characterized by pronounced aneuploidy.
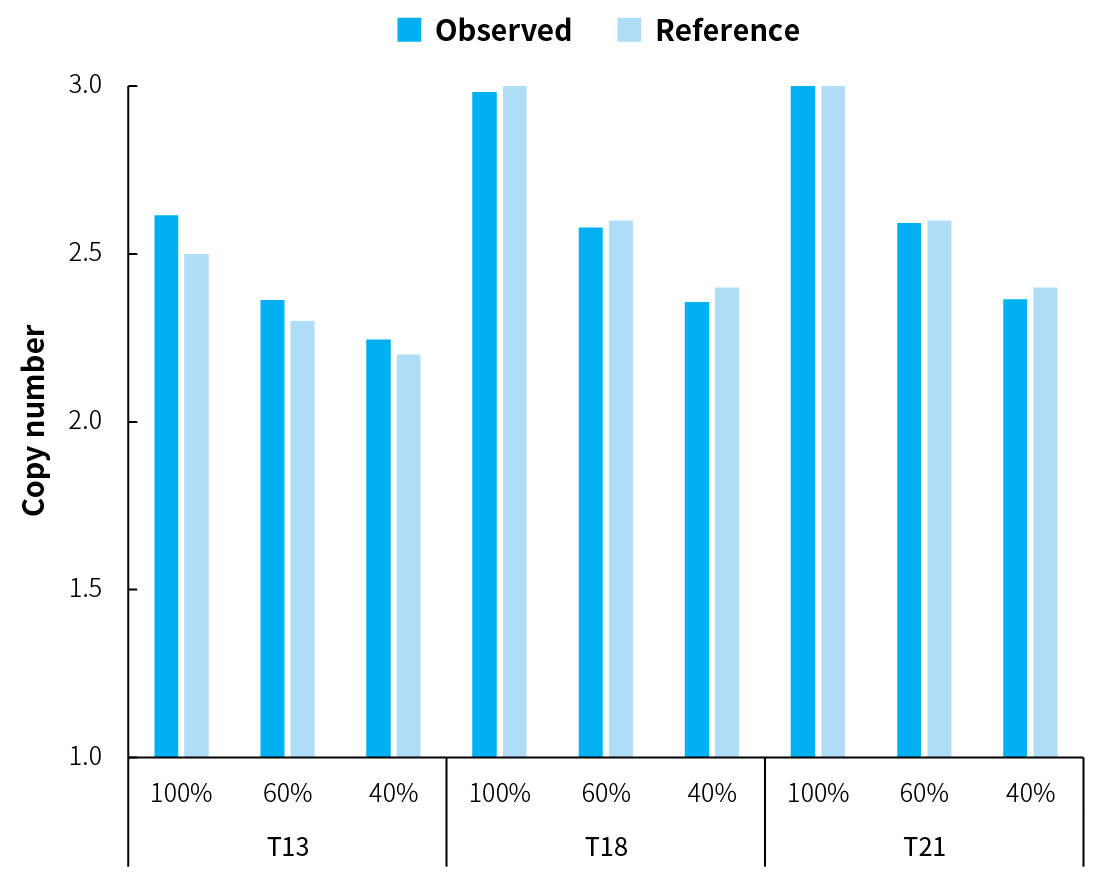
Figure 2. Analysis using the Nanodigmbio’s solution demonstrates concordance with previously characterized aneuploidies in defined mixtures. 30 pg of gDNA were amplified using NadPrep Single Cell WGA Kit. 100 ng of each amplified DNA were used for library preparation with NadPrep EZ DNA Library Preparation Module v2 and NadPrep Universal Stubby Adapter (UDI) Module. Libraries were sequenced on Illumina NovaSeq 6000 (PE150), generating approximately 1.5 Gb of raw data per sample. Variant analysis was performed with CNVkit in 100-kb bin sizes.
Note: Positive samples were cell line-derived gDNA reference standards characterized by pronounced trisomy 13/18/21 (LDT Bioscience, C2337T/C2338T/C2339T). Negative samples were Human Genomic DNA (Promega, G1471) known as euploidy. Defined mixtures were generated by mixing two gDNA standards (C2337T/C2338T/C2339T and G1471) at the indicated ratios, containing aneuploidies at varying proportions across different genomic locations.
For research use only. Not for use in diagnostic procedures.
Click here for more details on the "End-to-End Solution for PGT-A by LP-WGS".| #Package | Cap Color | Component |
Volume 1300101 (24 rxn) |
Volume 1300102 (96 rxn) |
| Box 1 |
|
Cell Lysis Buffer | 130 μL | 520 μL |
|
|
Cell Lysis Enzyme | 16 μL | 65 μL | |
|
|
1st Amplification Buffer | 320 μL | 1,270 μL | |
|
|
1st Amplification Enzyme | 32 μL | 130 μL | |
|
|
2nd Amplification Buffer | 810 μL | 2×1,615 μL | |
|
|
2nd Amplification Enzyme | 65 μL | 250 μL | |
|
/ |
NadPrep SP Beads |
2.5 mL |
8 mL |
Cell loss during isolation: if cells are lost during isolation or transfer, inspect and optimize the cell isolation and collection workflow. Recollect samples and confirm that each reaction tube contains a cell before proceeding.
Insufficient purity of gDNA: residual wash buffers, ethanol or other contaminants remaining after purification can inhibit the amplification reaction. Ensure ethanol is fully evaporated after purification, resuspend/dilute purified gDNA in Nuclease Free Water, and verify accurate quantification of the input samples.
I. If the initial input samples are cells: apoptosis of cells prior to lysis can lead to DNA degradation, which may produce allelic loss or chromosomal abnormalities. It is recommended to assess cell viability and proceed with lysis and amplification immediately when the cell status is acceptable. If immediate amplification is not possible, store lysed cells at -25 ~ -15℃ for ≤ 7 days to preserve genomic integrity.
II. If the initial input samples are bulk DNA: partial degradation of the input gDNA can likewise cause such artifacts. Use intact, non-degraded gDNA, or increase the initial input amount as appropriate.
Exogenous DNA contamination: this kit is highly sensitive to trace DNA; ultra-low amounts of contaminating DNA can yield microgram-level amplification products. Perform all procedures in a clean, DNA-free environment (e.g., PCR-clean area or laminar-flow hood), and use nuclease-free, DNA-free consumables and reagents. Thoroughly decontaminate work surfaces, pipettes and equipment, and include and closely monitor negative controls throughout the workflow to rapidly identify and trace contamination sources.
| Type | Product | Scale | Catalog# |
|
Single Cell WGA |
NadPrep Single Cell WGA Kit, 24 rxn | 24 rxn | 1300101 |
| NadPrep Single Cell WGA Kit, 96 rxn | 96 rxn | 1300102 |
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)

Nanodigmbio will contact you within 1 day.
If there are any problems, please contact us by 400 8717 699 / support@njnad.com