Reducing costs increases efficiency! It’s Indispensable!
View: 4436 / Time: 2024-09-06
01 Background
With the continuous innovation of gene sequencing technology, sequencing costs have significantly decreased while throughput has also dramatically increased. The Illumina NovaSeq 6000 S4 sequencing chip can produce over 2,400 Gb of data in just 44 hours, while the newly launched NovaSeq X Plus system can produce up to 16,000 Gb of data per dual flow cell run.
 Figure 1. Illumina NovaSeq X series and performance parameters.
Figure 1. Illumina NovaSeq X series and performance parameters.
Adapters are an integral and critical component of high-throughput sequencing libraries, with the Index sequences they contain used to differentiate between samples, allowing pooled libraries to be sequenced in a single run or within single flow cell lane. With the increase of sequencing throughput, the number of samples that can be processed per run has surged, but most Index combinations currently available on the market are insufficient to meet the actual demand.
Table 1. The number of Indexes required for different applications to meet specific sequencing depth or raw data.

* WGS: 100 Gb/sample, 30x;
** WES Panel: 15 Gb/sample, 100x;
Given issues such as Index Hopping, sample cross-contamination, and background noise[1-3] that can arise during library preparation, sequencing, and data analysis, several requirements have been proposed for Index design. On the one hand, the selection of Index length should must strike a balance between saving space for sample sequencing and increasing error tolerance[4]. On the other hand, Index sequences should be substantially different from one another to prevent cross-contamination of samples caused by Index Hopping. In addition, the Index should contain some code to quickly determine the accuracy of the test. Finally, Index designs must be flexible to satisfy biological requirements that could be imposed depending on the nuances of the application: such as GC content, sequence redundancy, presence of palindromes etc[4].
Theoretically, when any design requirements are met, such as minimum edit distance ≥ 3, homopolymers ≤ 2, 40%< GC < 60%, and no perfect self-complementarity, the number of permissible Index combinations is limited by Index length. For example, with a sequence length of 6 nt, there are at most 61 possible Index options; at 8 nt, this increases to 531 options; and with a sequence length of 10 nt, the number rises to 7,198, significantly improve Index diversity[5].
Table 2. Counts of four to 10 nucleotide, ≥ 3 edit distance sequence Indexes sets designed using EDITTAG.
 NadPrep Universal Stubby Adapter (UDI) Module series from Nanodigmbio, with its newly designed unique dual Index (10 nt), it can effectively addresses the increased Index demand brought about by higher sequencing throughput and helps solve the Index hopping issues in multiplexed sequencing. These adapters are compatible with the current mainstream 8 nt Index reading mode on the Illumina platforms, but also support 10 nt Index reading mode, to meet the needs of more samples in different application scenarios.
NadPrep Universal Stubby Adapter (UDI) Module series from Nanodigmbio, with its newly designed unique dual Index (10 nt), it can effectively addresses the increased Index demand brought about by higher sequencing throughput and helps solve the Index hopping issues in multiplexed sequencing. These adapters are compatible with the current mainstream 8 nt Index reading mode on the Illumina platforms, but also support 10 nt Index reading mode, to meet the needs of more samples in different application scenarios.
03 Exclusive Patent Design of Nanodigmbio (Patent No. CN 113999893B)
With optimized design, NadPrep Universal Stubby Adapter (UDI) Module has a unique dual 10 nt Index (barcode). It can support both 8 nt or 10 nt Index reading mode on the Illumina platforms, and comply with Illumina’s 8 nt 4-Index minimum color balance, easier to achieve multiplexed sequencing of pooled libraries.

Figure 2. NadPrep Universal Stubby Adapter (UDI) Module consists of universal stubby adapter and UDI Index Primer.
Table 3. Design principles of UDI Index Primer.

* The Index sequence strictly guarantees 3 edit distances even when 1 base insertion/indel.
04 Performance
4.1 Advantages of base balance design
NadPrep Universal Stubby Adapter (UDI) Module adopts the design concept of adapters with unique dual index from Nanodigmbio, designing for 4-Index balanced group: when ≥ 4 consecutive indexed libraries are mixed in equal ratio, the bases within the index achieve balance, effectively improving data separation and usage. In contrast, multi-index design solutions for Illumina platforms, such as Vendor I*'s UDI Adapter, may not meet sequencing requirements even with up to 12 consecutive indexes.

Figure 3. The base balance design of NadPrep Universal Stubby Adapter (UDI).
4.2 Uniform library yield and data output


Figure 5. Analysis of data cross contamination because of Index hopping. WGS libraries with 288 Indexes in NadPrep Universal Stubby Adapter (UDI) Module were mixed in a single lane for sequencing.
Note: Green indicates perfect match pairing barcode perfect match pairing barcode percentage > 98.5. A & B are two different batches.
4.4 Higher library yield
Using NadPrep DNA Library Preparation Kit (for Illumina®) coupled with NadPrep Universal Stubby Adapter (UDI) Module and NadPrep UDI Adapter Kit (for Illumina®) to prepare the library respectively. NadPrep Universal Stubby Adapter (UDI) has higher amplification efficiency and more efficient and stable library yield under the same number of cycles with the same input.

Figure 6. Library yield with different types of adapters. Yield of libraries prepared with different gDNA input amount by using NadPrep DNA Library Preparation Kit (for Illumina®) coupled with NadPrep Universal Stubby Adapter (UDI) Module and NadPrep UDI Adapter Module (for Illumina®) respectively.
Note: The sample is Human Genomic DNA (Promega, G1521).
4.5 Compatible with hybrid capture reagents
Pre-libraries prepared by the NadPrep Universal Stubby Adapter (UDI) Module are compatible with NadPrep Hybrid Capture Reagents and NadPrep NanoBlockers (for Illumina®️). The captured performance were comparable to the pre-libraries prepared by the NadPrep UDI Adapter (for Illumina®) (8 nt).

Figure 7. NadPrep Universal Stubby Adapter (UDI) is compatible with hybridization capture reagents. Human Genomic DNA (Promega, G1471) were prepared libraries by using NadPrep DNA Library Preparation Kit (for Illumina®) coupled with NadPrep UDI Adapter Kit (for Illumina®) and NadPrep Universal Stubby Adapter (UDI) Module respectively, completed hybrid capture with LungCancer Panel v1.0 (Nanodigmbio, 1001921).
Reference
[1] https://www.illumina.com.cn/content/dam/illuminamarketing/documents/products/whitepapers/index-hopping-white-paper-770-2017-004.pdf?linkId=36607862.
[2] Gilles A, Meglécz E, Pech N, et al. Accuracy and quality assessment of 454 GS-FLX Titanium pyrosequencing[J]. BMC genomics, 2011, 12: 1-11.
[3] Nguyen P, Ma J, Pei D, et al. Identification of errors introduced during high throughput sequencing of the T cell receptor repertoire[J]. BMC genomics, 2011, 12: 1-13.
[4] Bystrykh L V. Generalized DNA barcode design based on Hamming codes[J]. PloS one, 2012, 7(5): e36852.
[5] Faircloth B C, Glenn T C. Not all sequence tags are created equal: designing and validating sequence identification tags robust to indels[J]. PloS one, 2012,7(8): e42543.
With the continuous innovation of gene sequencing technology, sequencing costs have significantly decreased while throughput has also dramatically increased. The Illumina NovaSeq 6000 S4 sequencing chip can produce over 2,400 Gb of data in just 44 hours, while the newly launched NovaSeq X Plus system can produce up to 16,000 Gb of data per dual flow cell run.
 Figure 1. Illumina NovaSeq X series and performance parameters.
Figure 1. Illumina NovaSeq X series and performance parameters.Adapters are an integral and critical component of high-throughput sequencing libraries, with the Index sequences they contain used to differentiate between samples, allowing pooled libraries to be sequenced in a single run or within single flow cell lane. With the increase of sequencing throughput, the number of samples that can be processed per run has surged, but most Index combinations currently available on the market are insufficient to meet the actual demand.
Table 1. The number of Indexes required for different applications to meet specific sequencing depth or raw data.

* WGS: 100 Gb/sample, 30x;
** WES Panel: 15 Gb/sample, 100x;
*** Pan-cancer Panel: 5 Gb/sample, 1,000x;
Given issues such as Index Hopping, sample cross-contamination, and background noise[1-3] that can arise during library preparation, sequencing, and data analysis, several requirements have been proposed for Index design. On the one hand, the selection of Index length should must strike a balance between saving space for sample sequencing and increasing error tolerance[4]. On the other hand, Index sequences should be substantially different from one another to prevent cross-contamination of samples caused by Index Hopping. In addition, the Index should contain some code to quickly determine the accuracy of the test. Finally, Index designs must be flexible to satisfy biological requirements that could be imposed depending on the nuances of the application: such as GC content, sequence redundancy, presence of palindromes etc[4].
Theoretically, when any design requirements are met, such as minimum edit distance ≥ 3, homopolymers ≤ 2, 40%< GC < 60%, and no perfect self-complementarity, the number of permissible Index combinations is limited by Index length. For example, with a sequence length of 6 nt, there are at most 61 possible Index options; at 8 nt, this increases to 531 options; and with a sequence length of 10 nt, the number rises to 7,198, significantly improve Index diversity[5].
Table 2. Counts of four to 10 nucleotide, ≥ 3 edit distance sequence Indexes sets designed using EDITTAG.
 NadPrep Universal Stubby Adapter (UDI) Module series from Nanodigmbio, with its newly designed unique dual Index (10 nt), it can effectively addresses the increased Index demand brought about by higher sequencing throughput and helps solve the Index hopping issues in multiplexed sequencing. These adapters are compatible with the current mainstream 8 nt Index reading mode on the Illumina platforms, but also support 10 nt Index reading mode, to meet the needs of more samples in different application scenarios.
NadPrep Universal Stubby Adapter (UDI) Module series from Nanodigmbio, with its newly designed unique dual Index (10 nt), it can effectively addresses the increased Index demand brought about by higher sequencing throughput and helps solve the Index hopping issues in multiplexed sequencing. These adapters are compatible with the current mainstream 8 nt Index reading mode on the Illumina platforms, but also support 10 nt Index reading mode, to meet the needs of more samples in different application scenarios.03 Exclusive Patent Design of Nanodigmbio (Patent No. CN 113999893B)
With optimized design, NadPrep Universal Stubby Adapter (UDI) Module has a unique dual 10 nt Index (barcode). It can support both 8 nt or 10 nt Index reading mode on the Illumina platforms, and comply with Illumina’s 8 nt 4-Index minimum color balance, easier to achieve multiplexed sequencing of pooled libraries.

Figure 2. NadPrep Universal Stubby Adapter (UDI) Module consists of universal stubby adapter and UDI Index Primer.
Table 3. Design principles of UDI Index Primer.

* The Index sequence strictly guarantees 3 edit distances even when 1 base insertion/indel.
04 Performance
4.1 Advantages of base balance design
NadPrep Universal Stubby Adapter (UDI) Module adopts the design concept of adapters with unique dual index from Nanodigmbio, designing for 4-Index balanced group: when ≥ 4 consecutive indexed libraries are mixed in equal ratio, the bases within the index achieve balance, effectively improving data separation and usage. In contrast, multi-index design solutions for Illumina platforms, such as Vendor I*'s UDI Adapter, may not meet sequencing requirements even with up to 12 consecutive indexes.

Figure 3. The base balance design of NadPrep Universal Stubby Adapter (UDI).
4.2 Uniform library yield and data output

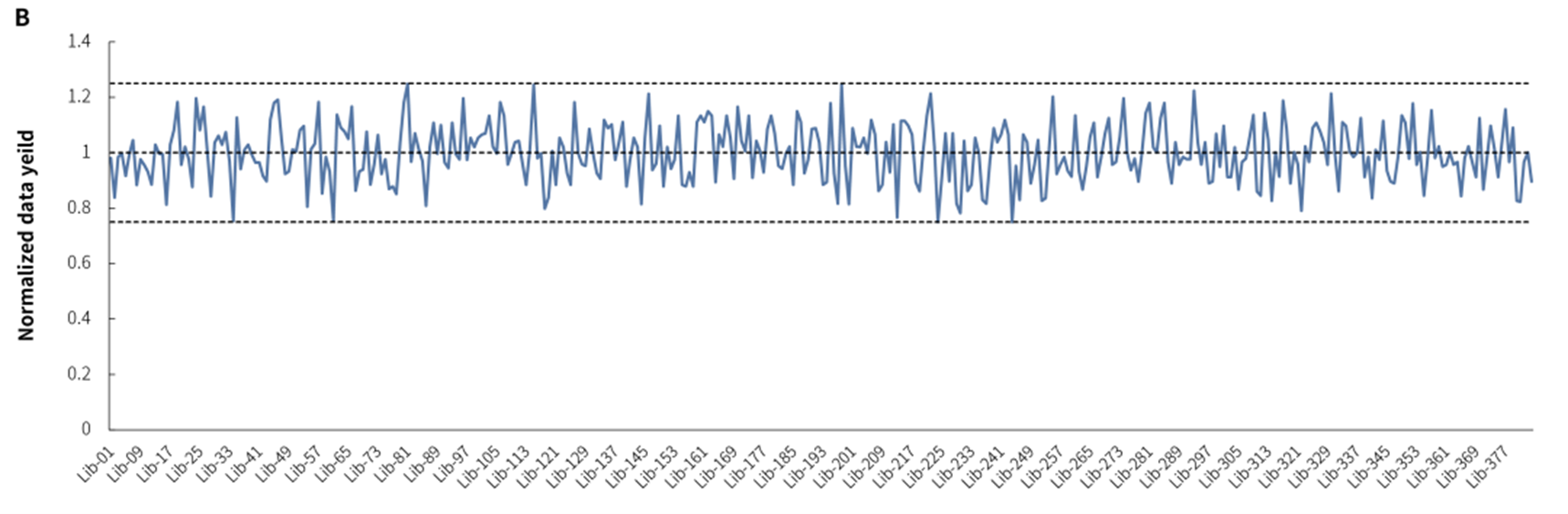
Figure 4. Application example of NadPrep Universal Stubby Adapter (UDI).A. Yield of libraries with 384 Indexes in NadPrep Universal Stubby Adapter (UDI) Module. According to the user manual of NadPrep DNA Library Preparation Kit (for Illumina®), the 10 ng fragmented gDNA were prepared libraries for 9 cycles with Index 1-384, respectively, resulting in an average library yield of > 10,00 ng. B. Effective splitting rate for libraries with 384 Indexes on Illumina platform. Sequencing mode: Novaseq 6000, PE150.
4.3 Effectively improve data accuracy
Figure 5. Analysis of data cross contamination because of Index hopping. WGS libraries with 288 Indexes in NadPrep Universal Stubby Adapter (UDI) Module were mixed in a single lane for sequencing.
Note: Green indicates perfect match pairing barcode perfect match pairing barcode percentage > 98.5. A & B are two different batches.
4.4 Higher library yield
Using NadPrep DNA Library Preparation Kit (for Illumina®) coupled with NadPrep Universal Stubby Adapter (UDI) Module and NadPrep UDI Adapter Kit (for Illumina®) to prepare the library respectively. NadPrep Universal Stubby Adapter (UDI) has higher amplification efficiency and more efficient and stable library yield under the same number of cycles with the same input.

Figure 6. Library yield with different types of adapters. Yield of libraries prepared with different gDNA input amount by using NadPrep DNA Library Preparation Kit (for Illumina®) coupled with NadPrep Universal Stubby Adapter (UDI) Module and NadPrep UDI Adapter Module (for Illumina®) respectively.
Note: The sample is Human Genomic DNA (Promega, G1521).
4.5 Compatible with hybrid capture reagents
Pre-libraries prepared by the NadPrep Universal Stubby Adapter (UDI) Module are compatible with NadPrep Hybrid Capture Reagents and NadPrep NanoBlockers (for Illumina®️). The captured performance were comparable to the pre-libraries prepared by the NadPrep UDI Adapter (for Illumina®) (8 nt).

Figure 7. NadPrep Universal Stubby Adapter (UDI) is compatible with hybridization capture reagents. Human Genomic DNA (Promega, G1471) were prepared libraries by using NadPrep DNA Library Preparation Kit (for Illumina®) coupled with NadPrep UDI Adapter Kit (for Illumina®) and NadPrep Universal Stubby Adapter (UDI) Module respectively, completed hybrid capture with LungCancer Panel v1.0 (Nanodigmbio, 1001921).
Reference
[1] https://www.illumina.com.cn/content/dam/illuminamarketing/documents/products/whitepapers/index-hopping-white-paper-770-2017-004.pdf?linkId=36607862.
[2] Gilles A, Meglécz E, Pech N, et al. Accuracy and quality assessment of 454 GS-FLX Titanium pyrosequencing[J]. BMC genomics, 2011, 12: 1-11.
[3] Nguyen P, Ma J, Pei D, et al. Identification of errors introduced during high throughput sequencing of the T cell receptor repertoire[J]. BMC genomics, 2011, 12: 1-13.
[4] Bystrykh L V. Generalized DNA barcode design based on Hamming codes[J]. PloS one, 2012, 7(5): e36852.
[5] Faircloth B C, Glenn T C. Not all sequence tags are created equal: designing and validating sequence identification tags robust to indels[J]. PloS one, 2012,7(8): e42543.
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)



