Detection and Surveillance of SARS-CoV-2 Variant Strains in Clinical Samples
View: 4992 / Time: 2024-08-28
On January 2, 2020, two patients with an unusual and severe form of pneumonia were identified in Wuhan, China. Researchers used metagenomic next-generation sequencing (mNGS) technology to decode the genome of the novel coronavirus within 5 days, after filtering out 98.5% and 99.38% of non-coronavirus gene sequences from the total sequencing data (with the coronavirus genetic sequences accounting for 1.5% and 0.62% of total RNA, respectively) (see Figure 1)[1]. Determining the full genome sequence of the virus, typing the virus, exploring the evolutionary spectrum of the virus, and closely monitoring the variation of the novel coronavirus are of great significance for precision prevention and control[2]. Although mNGS technology has obvious advantages in the detection of unknown pathogens, the inability to enrich the target region in sequencing results in a large proportion of non-target genomic sequencing data, leading to extremely high cost when detecting samples with low viral load.

Figure 1. Schematic diagram of the SARS-CoV-2 genome structure.
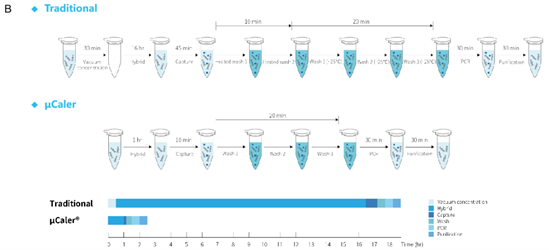
Targeted next-generation sequencing (tNGS) of pathogens can perfectly compensate for the shortcomings of mNGS. Traditional liquid-phase hybridization capture technology is a more efficient pathogen capture method in tNGS, but the disadvantage is that the hybridization time is too long and the process is slightly complicated. To solve this issue, we have developed a new liquid-phase hybridization capture technology called μCaler Hybrid System in 2022. The innovative design principle and mechanism (Figure 2. A) of this technology introduces a conjugation effect between capture probes, which improves sensitivity compared to traditional methods, while the entire hybridization capture time is shortened to 2.5 hours (Figure 2. B).
μCaler SARS-CoV-2 Panel that we recently launched aims to provide a simple, fast, and highly uniform SARS-CoV-2 whole genome sequencing solution. This panel is designed based on the original strain of the COVID-19 reference genome and can cover 29.9 Kb of the entire genome of the virus. In addition, the panel has been specially designed to enhance the capture ability of high-frequency mutation regions, such as the spike protein and RNA polymerase protein, which are particularly important for vaccine development.
Test 1: Detection of SARS-CoV-2 Cell Passage Virus (Omicron BA.1.1).
Sample type: RNA sample extracted from cell passage of SARS-Cov-2 variant strain (Omicron BA.1.1).
Using the reference genome as a template and the μCaler Hybrid system, which is a proprietary technology developed by Nanodigmbio, we designed the μCaler SARS-CoV-2 Panel to detect the whole genome sequence of cell passage viral strains. The on-target rate of all the three cell passage samples reached 95%, with 0.2x mean depth was greater than 99% (as shown in Figure 3. A). Giving the discontinuous replication mechanism of coronaviruses in the real samples, which generates a set of subgenomic RNAs (sgmRNAs) consistent with the virus genome sequence at the 5' end Leader sequence and 3' end sequence. Therefore, the coverage of whole genome shows deeper coverage at the 3' end of the genome (as shown in Figure 3. B), which also demonstrates that testing results of panel are very close to the biological characteristics of the virus.

Figure 3. Capture performance and genome-wide coverage of μCaler SARS-CoV-2 Panel on cell-passaged SARS-CoV-2 variants (Omicron BA.1.1). Libraries were prepared with RNA samples using the NadPrep Total RNA-To-DNA Module coupled with the NadPrep DNA Library Preparation Kit (for Illumina®), and a complete hybrid capture solution was developed using the μCaler SARS-CoV-2 Panel coupled with the μCaler Hybrid Capture Reagents and μCaler NanoBlockers (for Illumina®), with sequencing mode of MiniSeq SE 100. The sequencing data were analyzed using IRMA. A. Capture performance of the μCaler SARS-CoV-2 Panel on cell-passaged SARS-CoV-2; B. The genome-wide coverage of SARS-CoV-2.
Test 2: Whole-genome capture sequencing of SARS-CoV-2 in real samples.
Cell-passaged virus strains have higher virus abundance and higher genome integrity. The variation of pathogens in the real samples is complex, and there is a significant difference in virus load among different clinical samples. We selected four SARS-CoV-2 samples collected from oropharyngeal swabs for virus content assessment based on the virus sequence ORF genes and the N gene. The results showed that the CT values of the four samples were 24, 18, 16, and 19, respectively. Based on the on-target rate data, it is not difficult to see that even for real-world samples, the on-target rate can be consistently maintained above 80%(Figure 4).This also indicates that the μCaler hybrid capture system can perform stable capturing performance for real-world samples with different viral loads.
Finally, we used the IRMA software to assemble the virus's whole genome and used Nextclade for online analysis, completing the typing of four SARS-CoV-2 strains. We found that the first three samples tested carried the variant strain BF7, and the last sample carried the strain BA.5.2 (* data not shown). Therefore, from the above results, we can conclude that the μCaler whole-genome solution for the SARS-CoV-2 variant has the ability to detect the SARS-CoV-2 variant.

Figure 4. Capture performance and genome-wide coverage of μCaler SARS-CoV-2 Panel on oropharyngeal swab specimen collected. Libraries were prepared with RNA samples using the NadPrep Total RNA-To-DNA Module coupled with the NadPrep DNA Library Preparation Kit (for Illumina®), and a complete hybrid capture solution was developed using the μCaler SARS-CoV-2 Panel coupled with the μCaler Hybrid Capture Reagents and μCaler NanoBlockers (for Illumina®), with sequencing mode of MiniSeq SE 100. The sequencing data were analyzed using IRMA. A. Capture performance of the μCaler SARS-CoV-2 Panel on oropharyngeal swab specimen collected; B. The genome-wide coverage of SARS-CoV-2.
Reference
[1] Chen L, Liu W, Zhang Q, et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak[J]. Emerging microbes & infections, 2020, 9(1): 313-319.
[2] Qasem A, Shaw A M, Elkamel E, et al. Coronavirus disease 2019 (COVID-19) diagnostic tools: a focus on detection technologies and limitations[J]. Current issues in molecular biology, 2021, 43(2): 728-748.

Figure 1. Schematic diagram of the SARS-CoV-2 genome structure.
Targeted next-generation sequencing (tNGS) of pathogens can perfectly compensate for the shortcomings of mNGS. Traditional liquid-phase hybridization capture technology is a more efficient pathogen capture method in tNGS, but the disadvantage is that the hybridization time is too long and the process is slightly complicated. To solve this issue, we have developed a new liquid-phase hybridization capture technology called μCaler Hybrid System in 2022. The innovative design principle and mechanism (Figure 2. A) of this technology introduces a conjugation effect between capture probes, which improves sensitivity compared to traditional methods, while the entire hybridization capture time is shortened to 2.5 hours (Figure 2. B).

μCaler SARS-CoV-2 Panel that we recently launched aims to provide a simple, fast, and highly uniform SARS-CoV-2 whole genome sequencing solution. This panel is designed based on the original strain of the COVID-19 reference genome and can cover 29.9 Kb of the entire genome of the virus. In addition, the panel has been specially designed to enhance the capture ability of high-frequency mutation regions, such as the spike protein and RNA polymerase protein, which are particularly important for vaccine development.
μCaler ——Introduction of WGS Solution for the Variants of Coronavirus
In order to better verify the feasibility of μCaler - the total solution for coronavirus, we have collaborated with relevant institutions to test the specific performance of real samples.Test 1: Detection of SARS-CoV-2 Cell Passage Virus (Omicron BA.1.1).
Sample type: RNA sample extracted from cell passage of SARS-Cov-2 variant strain (Omicron BA.1.1).
Using the reference genome as a template and the μCaler Hybrid system, which is a proprietary technology developed by Nanodigmbio, we designed the μCaler SARS-CoV-2 Panel to detect the whole genome sequence of cell passage viral strains. The on-target rate of all the three cell passage samples reached 95%, with 0.2x mean depth was greater than 99% (as shown in Figure 3. A). Giving the discontinuous replication mechanism of coronaviruses in the real samples, which generates a set of subgenomic RNAs (sgmRNAs) consistent with the virus genome sequence at the 5' end Leader sequence and 3' end sequence. Therefore, the coverage of whole genome shows deeper coverage at the 3' end of the genome (as shown in Figure 3. B), which also demonstrates that testing results of panel are very close to the biological characteristics of the virus.

Figure 3. Capture performance and genome-wide coverage of μCaler SARS-CoV-2 Panel on cell-passaged SARS-CoV-2 variants (Omicron BA.1.1). Libraries were prepared with RNA samples using the NadPrep Total RNA-To-DNA Module coupled with the NadPrep DNA Library Preparation Kit (for Illumina®), and a complete hybrid capture solution was developed using the μCaler SARS-CoV-2 Panel coupled with the μCaler Hybrid Capture Reagents and μCaler NanoBlockers (for Illumina®), with sequencing mode of MiniSeq SE 100. The sequencing data were analyzed using IRMA. A. Capture performance of the μCaler SARS-CoV-2 Panel on cell-passaged SARS-CoV-2; B. The genome-wide coverage of SARS-CoV-2.
Test 2: Whole-genome capture sequencing of SARS-CoV-2 in real samples.
Cell-passaged virus strains have higher virus abundance and higher genome integrity. The variation of pathogens in the real samples is complex, and there is a significant difference in virus load among different clinical samples. We selected four SARS-CoV-2 samples collected from oropharyngeal swabs for virus content assessment based on the virus sequence ORF genes and the N gene. The results showed that the CT values of the four samples were 24, 18, 16, and 19, respectively. Based on the on-target rate data, it is not difficult to see that even for real-world samples, the on-target rate can be consistently maintained above 80%(Figure 4).This also indicates that the μCaler hybrid capture system can perform stable capturing performance for real-world samples with different viral loads.
Finally, we used the IRMA software to assemble the virus's whole genome and used Nextclade for online analysis, completing the typing of four SARS-CoV-2 strains. We found that the first three samples tested carried the variant strain BF7, and the last sample carried the strain BA.5.2 (* data not shown). Therefore, from the above results, we can conclude that the μCaler whole-genome solution for the SARS-CoV-2 variant has the ability to detect the SARS-CoV-2 variant.

Figure 4. Capture performance and genome-wide coverage of μCaler SARS-CoV-2 Panel on oropharyngeal swab specimen collected. Libraries were prepared with RNA samples using the NadPrep Total RNA-To-DNA Module coupled with the NadPrep DNA Library Preparation Kit (for Illumina®), and a complete hybrid capture solution was developed using the μCaler SARS-CoV-2 Panel coupled with the μCaler Hybrid Capture Reagents and μCaler NanoBlockers (for Illumina®), with sequencing mode of MiniSeq SE 100. The sequencing data were analyzed using IRMA. A. Capture performance of the μCaler SARS-CoV-2 Panel on oropharyngeal swab specimen collected; B. The genome-wide coverage of SARS-CoV-2.
Reference
[1] Chen L, Liu W, Zhang Q, et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak[J]. Emerging microbes & infections, 2020, 9(1): 313-319.
[2] Qasem A, Shaw A M, Elkamel E, et al. Coronavirus disease 2019 (COVID-19) diagnostic tools: a focus on detection technologies and limitations[J]. Current issues in molecular biology, 2021, 43(2): 728-748.
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)



