New Arrival | A more efficient comprehensive tNGS solution for RNA and DNA pathogen detection within the same library.
Infectious diseases caused by pathogenic microorganisms are among the significant causes leading high mortality and morbidity in humans. The types of pathogenic infections are diverse, including parasites, fungi, bacteria, mycoplasma/chlamydia/spirochetes/Rickettsia, viruses, etc. Specific diagnostic and therapeutic measures are often required for different pathogenic infections. Therefore, rapidly and accurately detecting the causative pathogen is crucial for the diagnosis, treatment, and management of infectious diseases.
RNA viruses are susceptible to being missed detection, and are commonly associated with mixed infections involving multiple systems. Among them, dual-pathogen mixed infections are commonly observed in the respiratory system, with the main patterns being viral-viral co-infections, viral-bacterial co-infections, and mycoplasma pneumoniae-viral co-infections, accounting for 15.8% to 31%. According to monitoring data on respiratory tract infections involving a total of 233,037 cases released by the Chinese Center for Disease Control and Prevention at the end of 2021, among the 8 viruses causing acute respiratory infections, the top 4 are all RNA viruses: Influenza Virus (IFV), Respiratory Syncytial Virus (RSV), Human Rhinovirus (HRV), and Human Parainfluenza Pirus (HPIV). So, is it necessary to conduct RNA & DNA co-detection? Who are the suitable candidates for RNA & DNA co-detection?
DNA detection process is suitable for suspected DNA pathogen infections, such as bacteria, fungi, parasites, DNA viruses, especially intracellular bacteria (e.g., Mycobacterium tuberculosis) and fungi with thicker cell walls (e.g., Cryptococcus). However, RNA detection process is applicable for suspected RNA virus infections, such as influenza virus, respiratory syncytial virus, coronaviruses, and other RNA viruses. When viral infection are challenging to exclude, simultaneous RNA and DNA co-detection is recommended.
However, the dual-process detection of RNA and DNA is costly, and there is an urgent need for an RNA & DNA single-tube library co-preparation scheme to reduce detection costs and shorten detection time. Combining targeted next-generation sequencing (tNGS) of pathogenic microorganisms further aids in rapid and accurate clinical diagnosis and treatment of infections.
02 RNA & DNA co-detection using targeted next-generation sequencing (tNGS) comprehensive solution.
2.1 Solution process
Nanodigmbio provides an Comprehensive tNGS Solution for pathogenic microorganisms detection, including: NadAuto-series Automated Workstations, NadPrep RNA & DNA Library Co-Preparation Module, various adapters suitable for dual sequencing platforms, NadPrep ES Hybrid Capture Reagents, Blockers, NEX-t Panel v1.0, online Bioinformatics Analysis Visual System named XCapViz and the report interpretation. The entire process of the solution takes only 14 hours, including Nucleic Acid Extraction (~1.5 hr), RNA & DNA Library Co-Preparation (~3 hr), Hybrid Capture with NEX-t Panel v1.0 (~4 hr), NGS sequencing (~5 hr), Bioinformatics Analysis Visual System named XCapViz, and automated report interpretation (~0.5 hr).

Figure 1. Comprehensive tNGS Solution for pathogenic microorganisms detection
2.2 Solution Features
① RNA & DNA Library Co-Preparation
• Single process implementation of RNA & DNA co-detection: Simultaneously detect RNA & DNA pathogens in one library preparation process to preventing omissions.
• Flexible compatibility with different sample types: Suitable for various clinical sample scenarios, flexibly compatible with different sample types.
② NEX-t Panel v1.0
• Broad coverage of pathogen spectrum in single detection: NEX-t Panel design covers 450+ genera of pathogens, meeting over 99% of clinical detection needs.
• Positive enrichment to enhance detection sensitivity: Eliminate host background interference and significantly improve the sensitivity of target pathogen detection.
• Enhance the ability to detect drug resistance/virulence genes: the probes in NEX-t Panel are design covers drug resistance and virulence-related genes, capturing variations deeply, and supporting precision diagnosis and treatment of clinical infections.
③ XCapViz ——Visual Bioinformatics Analysis Platform
• The XCapViz visual bioinformatics analysis platform offers intelligent and rapid bioinformatics analysis, generating detection reports with a single click.
• Data analysis and the processes are localized to ensure data security.
03 RNA & DNA Library Co-Preparation
3.1 Introduction
Nanodigmbio launched the targeted pathogen detection panel, NEX-t Panel v1.0, in September 2023. This panel targets viruses, bacteria, fungi, parasites, as well as housekeeping genes and drug-resistance related genes and more. By fully leveraging the fault-tolerant advantages of probe hybridization, the panel size has been streamlined to approximately 8,000 probes, maintaining enrichment information while reducing the complexity of implementation.
NadPrep RNA & DNA Library Co-Preparation Module is developed for high-throughput sequencing platforms. It enables efficient double-stranded DNA conversion of RNA samples, fragmentation of double-stranded DNA and DNA samples, end repair, and A-tailing in a single reaction system. It is suitable for detecting mixed pathogenic microorganism samples containing 10-100 ng RNA and 1-100 ng DNA. The pre-library prepared using this module is seamlessly conjunction with NEX-t Panel v1.0 and NadPrep ES Hybrid Capture Reagents provides a faster, more accurate, and more economical solution for tNGS detection.

Figure 2. Experimental workflow of the NadPrep RNA & DNA Library Co-Preparation Module
3.2 Features
 Figure 3. Product Features of the NadPrep RNA & DNA Library Co-Preparation Module
Figure 3. Product Features of the NadPrep RNA & DNA Library Co-Preparation Module04 Performance
4.1 Flexible Compatibility with Mixed Samples with Different RNA & DNA Ratios
In conventional pathogen detection workflows, it is typically necessary to separately prepare libraries for RNA and DNA in clinical samples. However, in scenarios involving precious samples, suspected mixed infections, or urgent testing requirements, multiple testing platforms and expert guidelines recommend the simultaneous detection of RNA and DNA, which is expected to significantly increase clinical benefits. We initiated library preparation using the NadPrep RNA & DNA Co-Preparation Module with mixed samples of different input amounts of K562 cell line RNA and human genomic DNA standard (Promega, G1521). The results demonstrate that efficient and stable library preparation and fragmentation can be achieved with different ratios and input amounts of RNA & DNA templates, whether on the Illumina or MGI platforms.
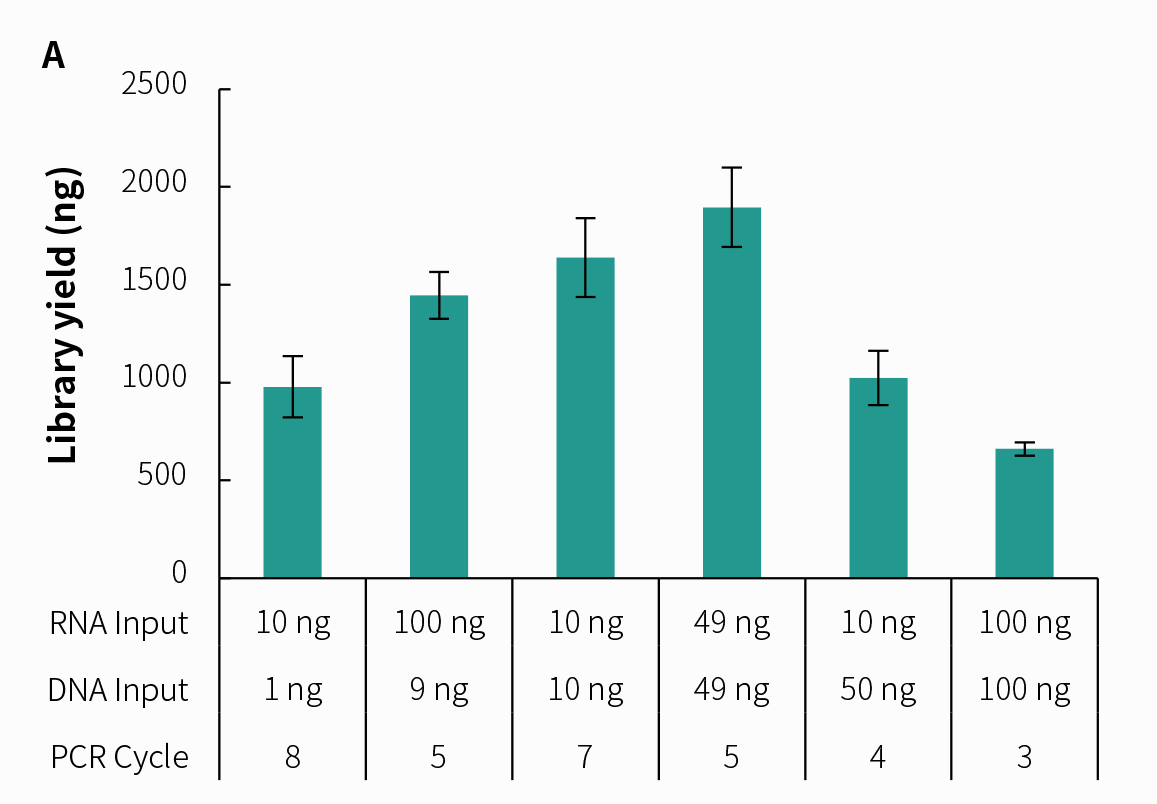

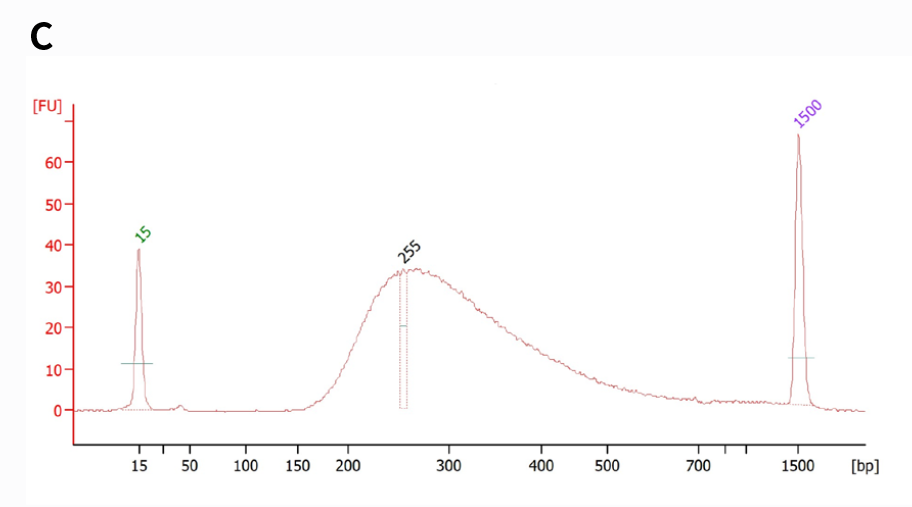
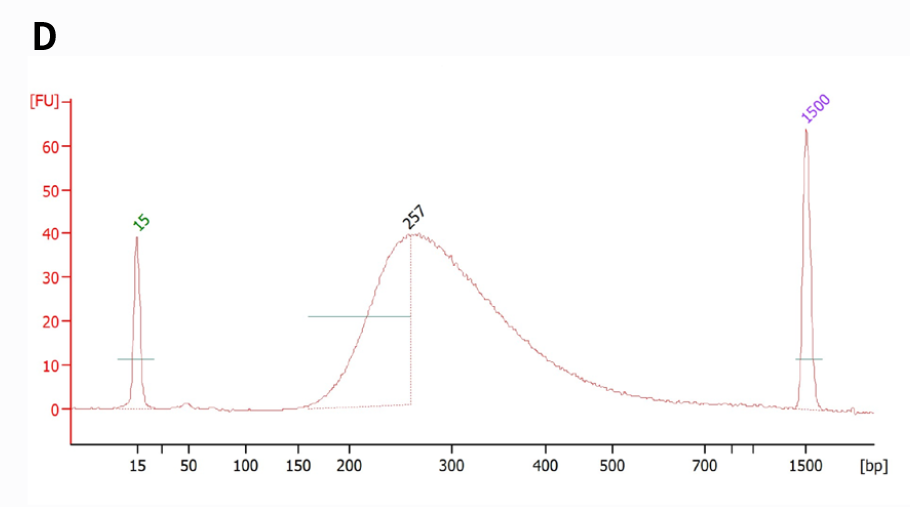
Figure 4. NadPrep RNA & DNA Library Co-Preparation Module for library co-preparation with mixed RNA & DNA samples in varying initial ratios. Utilize NadPrep RNA & DNA Library Co-Preparation Module in conjunction with NadPrep Universal Adapter (MDI) Module (for MGI) and NadPrep Universal Stubby Adapter (UDI) Module for library co-preparation. A & C. Library yield and size distribution (MDI); B & D. Library yield and size distribution (UDI).
4.2 Support for Dual Sequencing PlatformsDue to its higher sensitivity and lower cost in detection, tNGS technology has attracted widespread attention in clinical applications. We prepared pre-libraries using simulated pathogenic microorganism samples with the NadPrep RNA & DNA Library Co-Preparation Module, followed by hybrid capture using NadPrep ES Hybrid Capture Reagents and NEX-t Panel v1.0, with a hybridization time of 2 hours. The results demonstrate that libraries prepared using the NadPrep RNA & DNA Library Co-Preparation Module can effectively detect pathogenic microorganisms on both Illumina and MGI sequencing platforms.
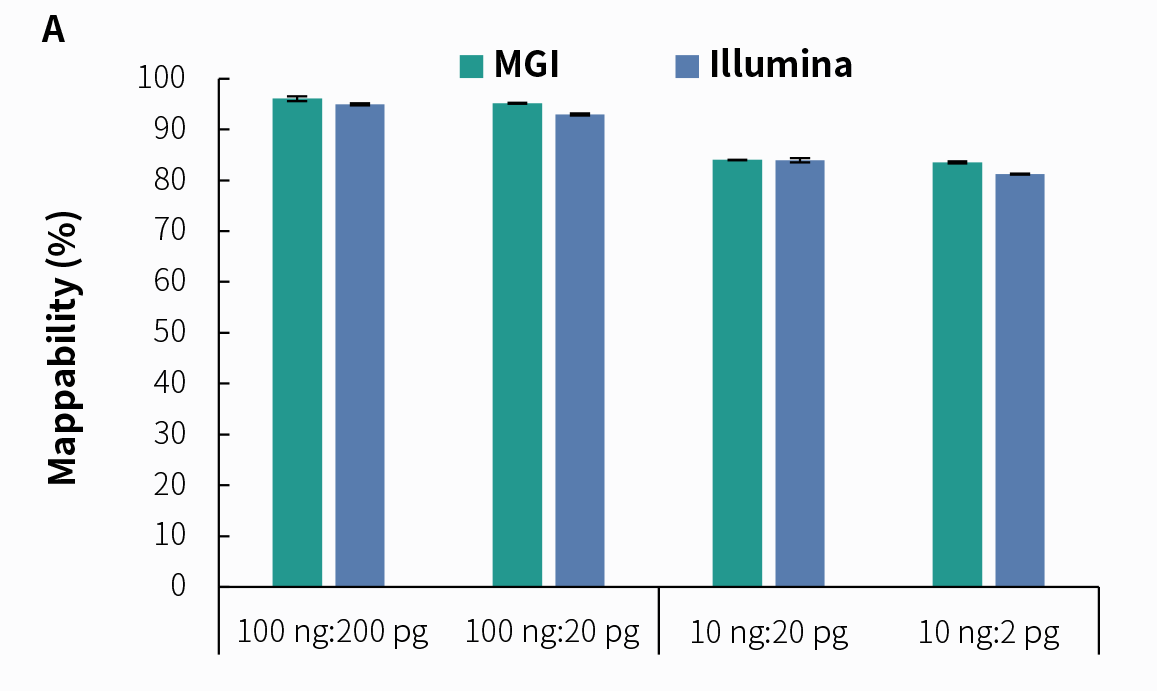

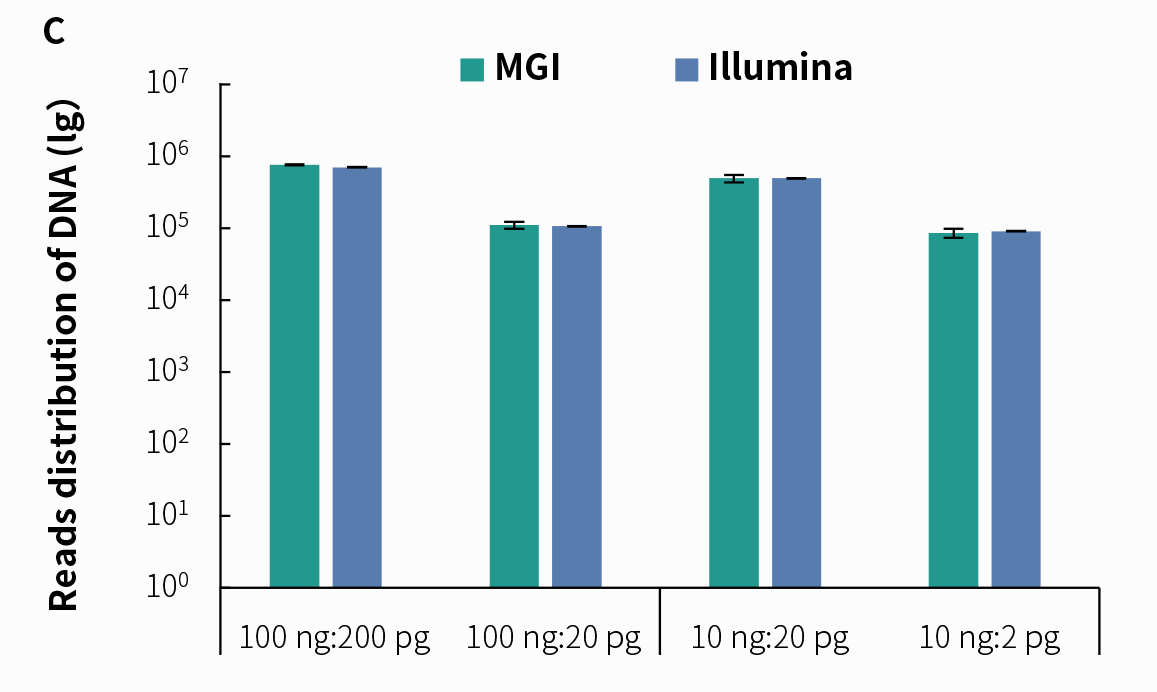
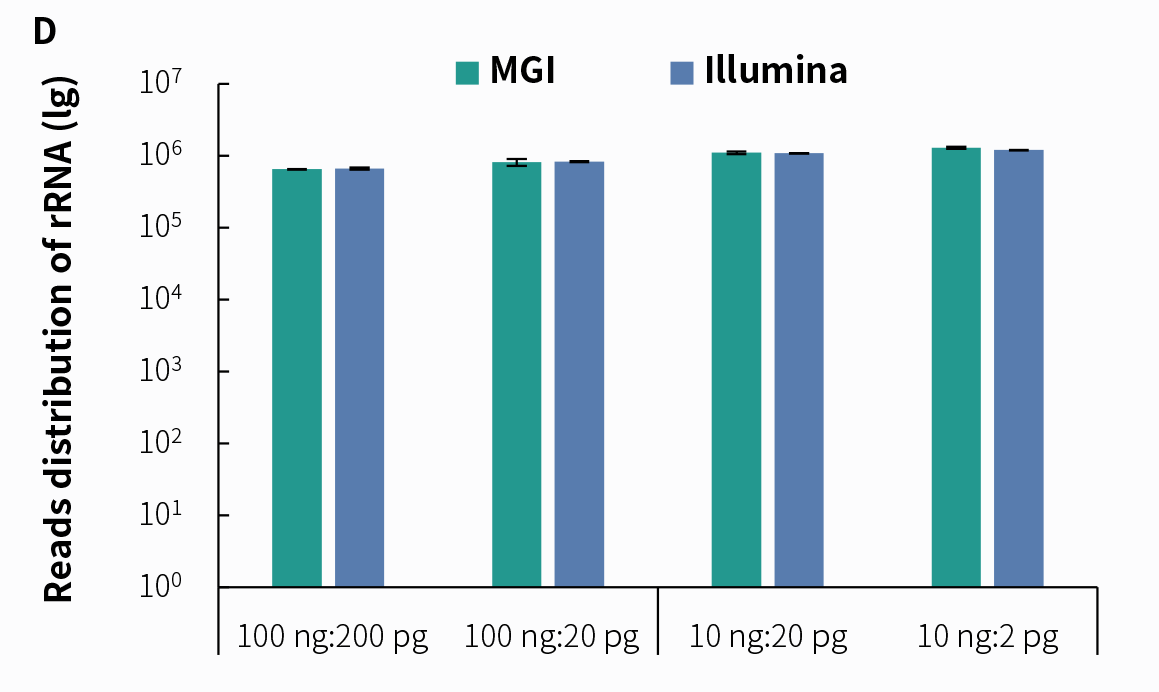
Figure 5. Detection of tNGS Total Solution with NadPrep RNA & DNA co-preparation on dual sequencing platforms. A. Mappability; B. Reads distribution of Coronavirus; C. Reads distribution of Candida albicans; D. Reads distribution of rRNA. Pre-libraries preparation were performed using NadPrep RNA & DNA Library Co-Preparation Module. Hybridization capture were completed with NadPrep ES Hybrid Capture Reagents and NEX-t Panel v1.0, followed by sequencing on DNBSEQ-T7, PE150 and Illumina NovaSeq 6000, PE150. For each sample, 0.5 Gb was used for data analysis.
Note: The scale on the X-axis represents the proportional initial input amounts of the host-mimicking mixed nucleic acid samples to the input amounts of the pathogenic mixed nucleic acid samples. The host-mimicking mixed nucleic acid samples consist of Human Brain Total RNA Standard (Clontech, 636530) and Human Genomic DNA Standard (Promega, G1521) mixed at a 1:1 ratio. The pathogenic mixed nucleic acid samples consist of the Reference material for SARS-CoV-2 Omicron Variant Genomic RNA (National Institute of Metrology, NIM-RM 5225; 1 pg equals 50 copies) and Candida albicans standard (ATCC, 10231; 1 pg equals 6000 copies) mixed at a 1:1 ratio.4.3 tNGS Effectively Enhances Detection Sensitivity
4.3.1 tNGS vs mNGS
In the diagnosis of infectious diseases, it is necessary to identify the causative pathogens quikly and accurately. However, while mNGS technology provides nearly comprehensive coverage of microbial species, its results are often complex to interpret, and its capability to detect low-load microorganisms is relatively weak. In contrast, tNGS technology offers advantages in relatively broad spectrum coverage, covering over 95% of common pathogens associated with clinical infections, and effectively enriching low-load pathogens. This addresses the current shortcomings in infectious disease detection, thereby enhancing the reliability of detection results. We conducted tests using the same sequencing raw data (0.5 Gb)with both tNGS and mNGS technologies for pathogen detection, and the results indicate that the tNGS approach effectively enriches information on low-load pathogenic microorganisms.
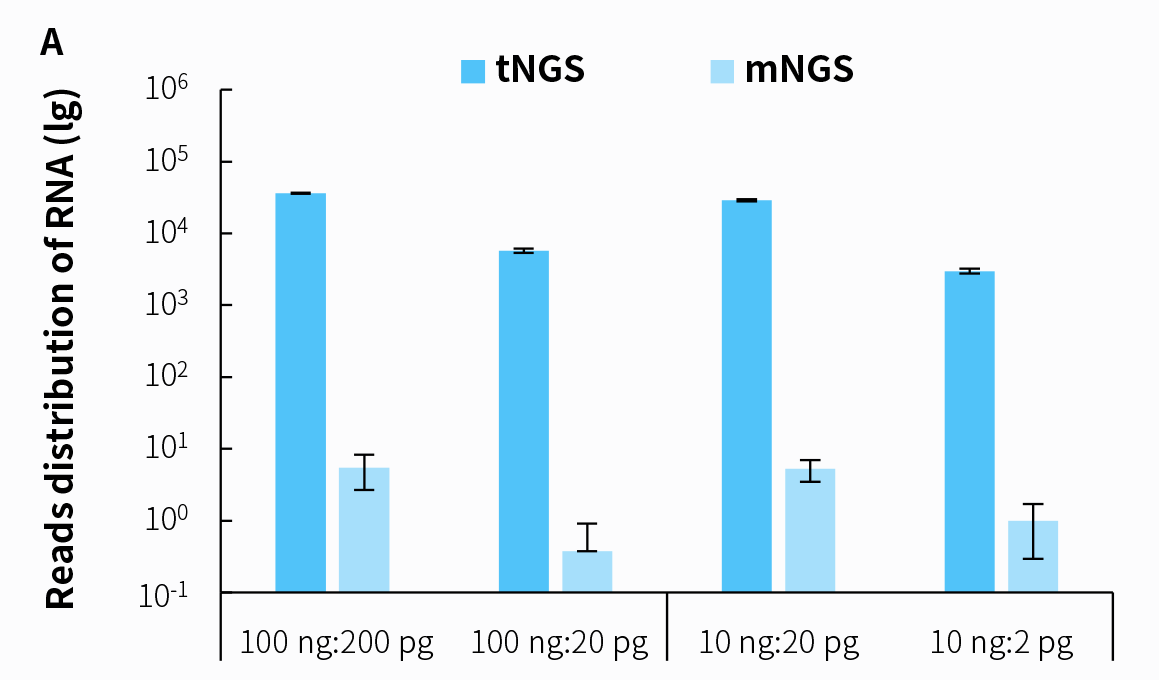

Figure 6. Distribution of detected reads in simulated pathogenic microbial samples using tNGS and mNGS based on the NadPrep RNA & DNA Library Co-Preparation Module. A. Reads distribution of Coronavirus; B. Reads distribution of Candida albicans.
4.3.2 Effective Improvement in Detection Sensitivity for RNA Virus
Another highly regarded aspect of RNA & DNA co-library preparation is its effectiveness in detecting RNA viruses. We conducted pre-library preparation using the NadPrep RNA & DNA Library Co-Preparation Module (Co-Prep) and the NadPrep Total RNA-To-DNA Module coupled with the NadPrep DNA Universal Library Preparation Kit (RNA-Prep) for the same simulated RNA virus samples. Hybrid capture was then performed using the NEX-t Panel v1.0. Analysis was conducted using the same amount of dataset. The results indicate that both library preparation methods yield consistent results for RNA virus detection.

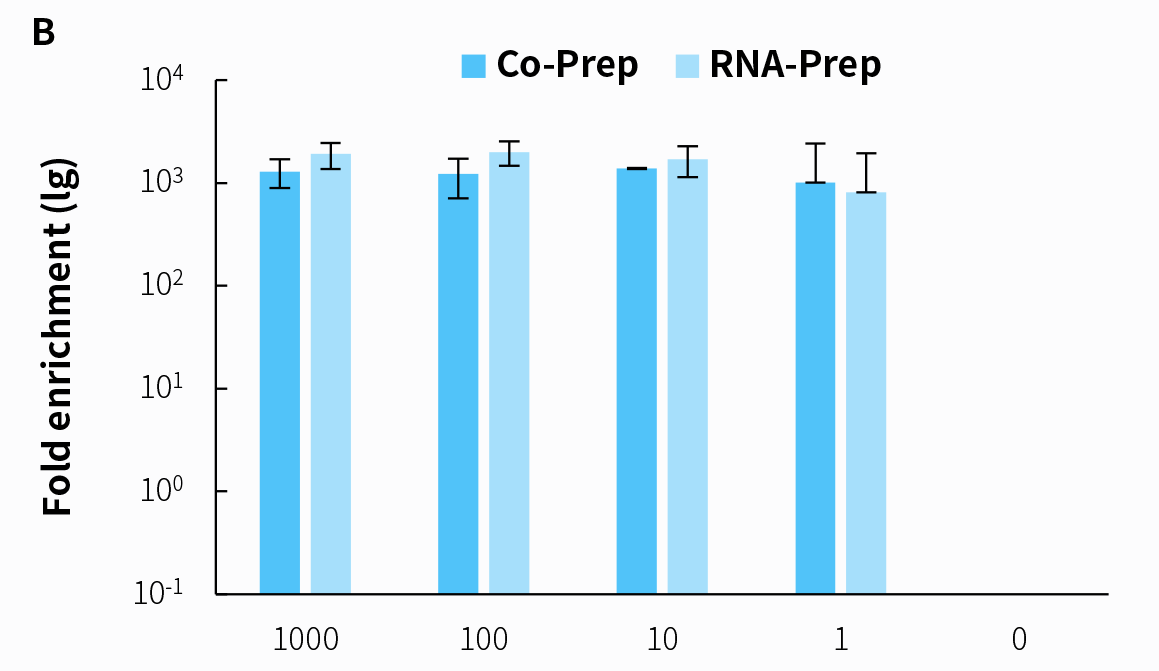
Figure 7. Capture performance of mixed RNA samples in varying copy numbers with co-preparation and RNA-preparation. A. Reads distribution; B. Enrichment fold.
Note: The X-axis represents the copy number of the Reference material for SARS-CoV-2 Omicron Variant Genomic RNA. The samples are derived from a mixture of Human Brain Total RNA Standard (Clontech, 636530) and Omicron virus samples in varying copy numbers.4.3.3 Effective Enhancement in Detection Sensitivity for DNA Pathogen
We conducted pre-library preparation of simulated DNA pathogen samples using both the NadPrep RNA & DNA Library Co-Preparation Module (Co-Prep) and the NadPrep EZ DNA Library Preparation Module v2 (DNA-Prep) separately, in conjunction with NadPrep Universal Stubby Adapter (UDI) Adapter for pre-library preparation, followed by hybrid capture using NadPrep ES Hybrid Capture Reagents and the NEX-t Panel v1.0. Comparing the deduplicated coverage depth obtained from the two library preparation methods with the same amount of dataset, the results showed consistent detection of DNA pathogens.

Figure 8. Captured coverage depth after duplication removal for samples with different microbial contents with co-preparation and DNA-preparation.
Note: Simulated microbial community samples of 0.001% - 1% MSA-1003 were created by diluting a mixture of 20 strains of genomic material (ATCC, MSA-1003) using the human genomic DNA standard (Promega, G1471) at various ratios. Sequencing platform: Illumina NovaSeq 6000, PE150.
4.4 Low GC Bias
We compared the detection performance of the NadPrep RNA & DNA Library Co-Preparation Module with those of other vendor A* and B* across microbial community samples with different GC content. The results indicate that the NadPrep® RNA & DNA Library Co-Preparation Module exhibits no significant GC bias.
 Figure 9. Uniform coverage of regions with various GC content using NadPrep RNA & DNA Library Co-Preparation Module. GC bias of whole-genome libraries for A. Staphylococcus aureus, B. Bacillus subtilis, and C. Lactobacillus fermentum with different GC content.
Figure 9. Uniform coverage of regions with various GC content using NadPrep RNA & DNA Library Co-Preparation Module. GC bias of whole-genome libraries for A. Staphylococcus aureus, B. Bacillus subtilis, and C. Lactobacillus fermentum with different GC content.4.5 Capture Performance of Clinical Sample
We compared the detection performance of the NadPrep RNA & DNA Library Co-Preparation Module with other vendor, labeled as A* and B*, using clinical nasal virus samples. The results indicate a clear sensitivity advantage of the NadPrep RNA & DNA Library Co-Preparation Module for nasal virus detection.
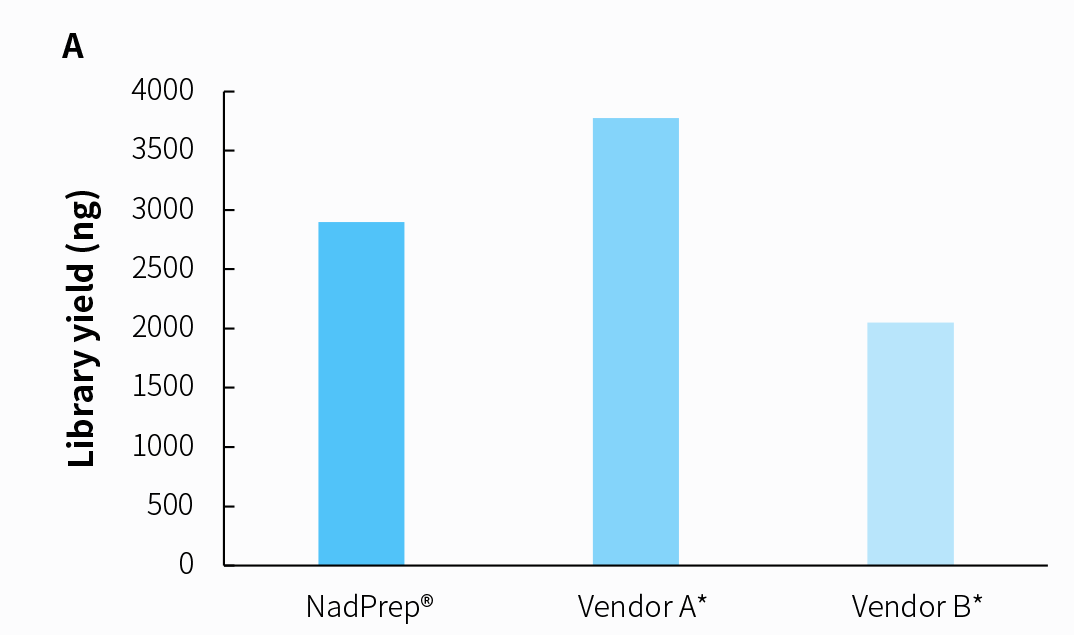

Figure 10. Detection performance of tNGS Total Solution for clinical samples with NadPrep RNA & DNA co-preparation. A. Library yield; B. Reads ratio of Rhinovirus A and Rhinovirus C.
05 Summary
With the official release of the NadPrep RNA & DNA Library Co-Preparation Module, combined with the efficient and streamlined NEX-t Panel v1.0, rapid and stable NadPrep ES Hybrid Capture Reagents, as well as the Bioinformatics Analysis Visual System named XCapViz and automated interpretation report, Nanodigmbio offers a Comprehensive tNGS Solution for pathogenic microorganisms detection. This solution not only reduces the cost and shortens the detection time significantly but also enhances the efficiency of detection. It provides robust support for the rapid and accurate diagnosis and treatment of clinical infections.
06 Solution
Product
Catalog#
Scale
Lib-Prep NadPrep RNA & DNA Library Co-Preparation Module,24 rxn
NadPrep RNA & DNA Library Co-Preparation Module,24 rxn
1002411
1002412
24 rxn
96 rxn
Adapter NadPrep Universal Stubby Adapter (UDI) Module series 1003240, ect 24/96/1152 rxn
NadPrep Universal Adapter (MDI) Module (for MGI) series 1003711, ect 24/96/1152 rxn
1003240, ect
1003711, ect
24/96/1152 rxn
24/96/1152 rxn
Hybrid Capture Reagent NadPrep ES Hybrid Capture Reagents, 16 rxn
NadPrep ES Hybrid Capture Reagents, 96 rxn
1005402
1005401
16 rxn
96 rxn
Panel NEX-t Panel v1.0, 16 rxn
NEX-t Panel v1.0, 96 rxn
1001972
1001971
16 rxn
96 rxn
Reference
[1] Li Z J, Zhang H Y, Ren L L, et al. Etiological and epidemiological features of acute respiratory infections in China[J]. Nature communications, 2021, 12(1): 5026.
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)



